Simulation method for CL-20/TNT (Hexanitrohexaazaisowurtzitane/Trinitrotoluene) eutectic crystal base compound
An analog method, the technology of CL-20, is applied in the direction of instrumentation, calculation, electrical digital data processing, etc., and can solve problems such as refractory degradation, and achieve good effect, low cost, and good visualization effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
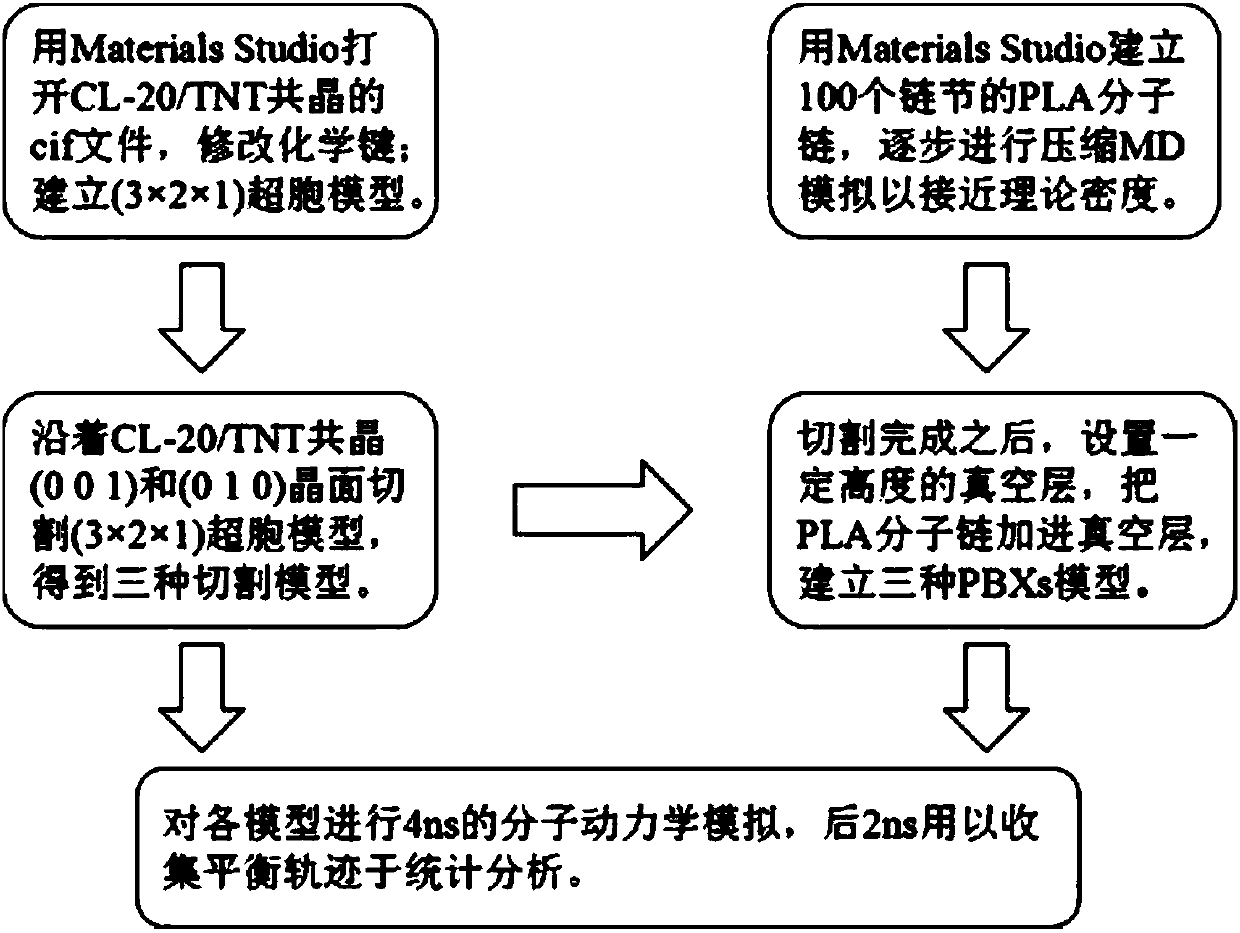
[0031] Establishment of cutting model and corresponding PBXs model
[0032] Step 1: Open the cif file of CL-20 / TNT eutectic with Materials Studio, change all the N-O single bonds in the file to virtual double bonds, and change the benzene ring to a resonant virtual double bond benzene ring;
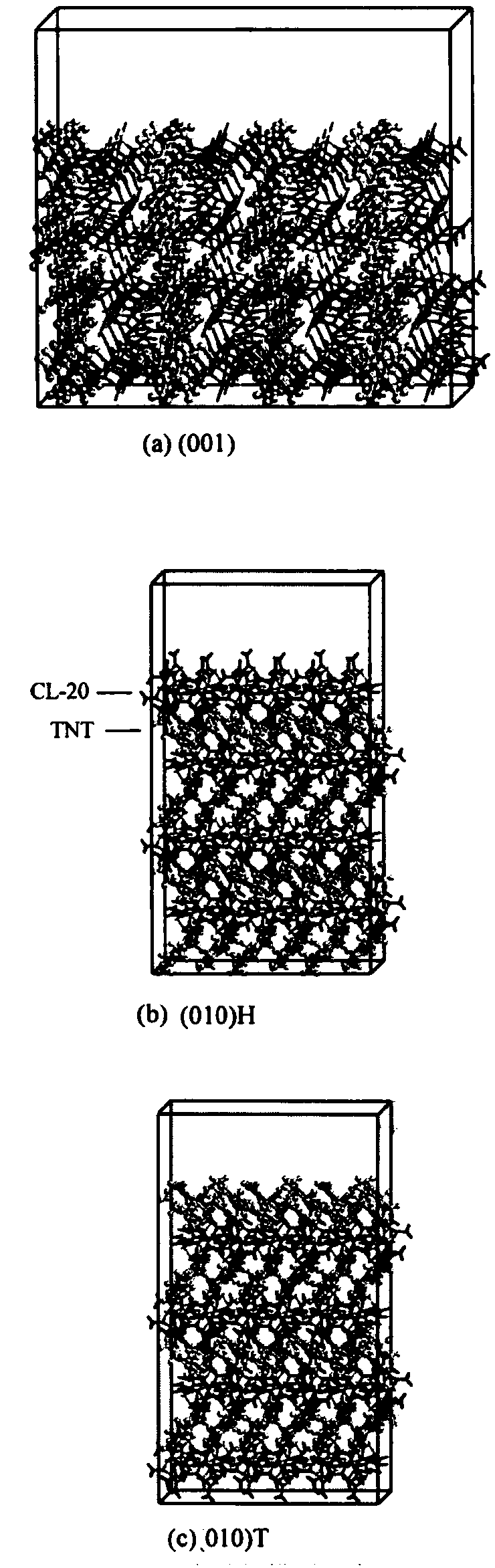
[0033] Step 2: Build a 3a×2b×1c supercell model, then click Build in the MS taskbar, select Surfaces, and then select Cleave Surface, then enter 0 0 1 in cleave plane, and click Cleave to build a cutting model;
[0034] Step 3: Establish the PLA molecular chain, select 100 chain links, place the constructed PLA molecular chain in a periodic box, gradually compress and perform molecular dynamics simulation until the density of the PLA molecular chain reaches the theoretical value;
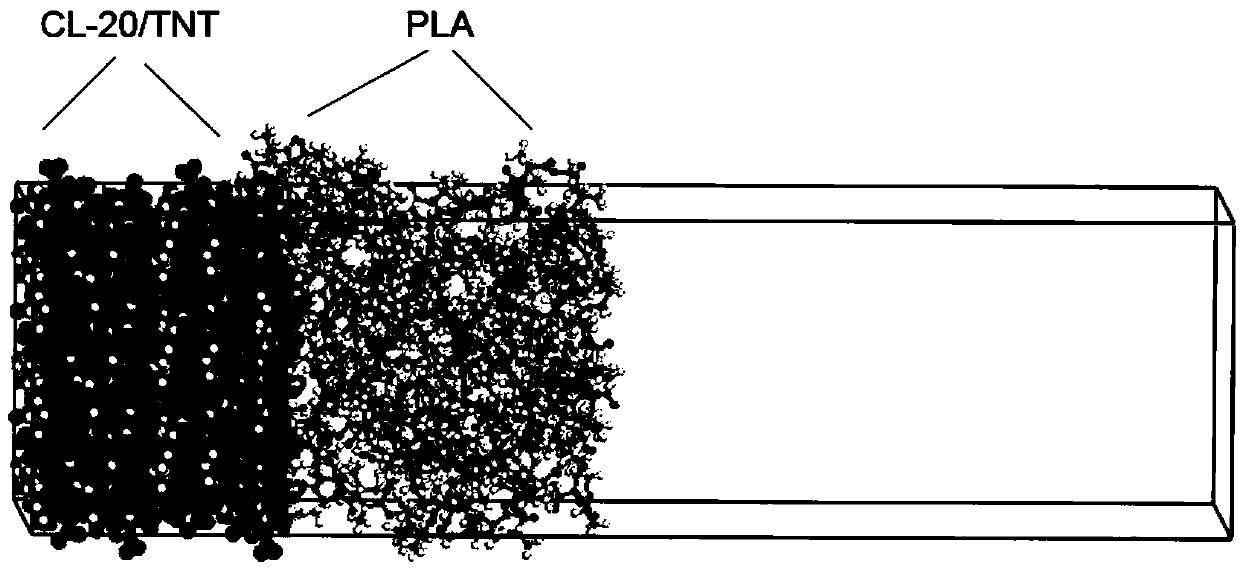
[0035] The fourth step: set a vacuum layer on the upper part of the cut model obtained in the second step, so that the size of the bonded model in the Z direction conforms to: Ztotal≈2×(Zcocrystal+Zpolymer). Z...
Embodiment 2
[0046] Establishment of cutting model and corresponding PBXs model
[0047] Step 1: Open the cif file of CL-20 / TNT eutectic with Materials Studio, change all the N-O single bonds in the file to virtual double bonds, and change the benzene ring to a resonant virtual double bond benzene ring;
[0048] Step 2: Build a 3a×2b×1c supercell model, then click Build in the MS task bar, select Surfaces, and then select Cleave Surface, then enter 0 1 0 in cleave plane, and set top in the Position section Angstrom value is Click Cleave to create a cutting model;
[0049] Step 3: Establish the PLA molecular chain, select 100 chain links, place the constructed PLA molecular chain in a periodic box, gradually compress and perform molecular dynamics simulation until the density of the PLA molecular chain reaches the theoretical value;
[0050] The fourth step: set a vacuum layer on the upper part of the cut model obtained in the second step, so that the size of the bonded model in the Z di...
Embodiment 3
[0054] Establishment of cutting model and corresponding PBXs model
[0055] Step 1: Open the cif file of CL-20 / TNT eutectic with Materials Studio, change all the N-O single bonds in the file to virtual double bonds, and change the benzene ring to a resonant virtual double bond benzene ring;
[0056] Step 2: Build a 3a×2b×1c supercell model, then click Build in the MS task bar, select Surfaces, and then select Cleave Surface, then enter 0 1 0 in cleave plane, and set top in the Position section Angstrom value is Click Cleave to create a cutting model;
[0057] Step 3: Establish the PLA molecular chain, select 100 chain links, place the constructed PLA molecular chain in a periodic box, gradually compress and perform molecular dynamics simulation until the density of the PLA molecular chain reaches the theoretical value;
[0058] The fourth step: set a vacuum layer on the upper part of the cut model obtained in the second step, so that the size of the bonded model in the Z di...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Spread coefficient | aaaaa | aaaaa |
| Spread coefficient | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com