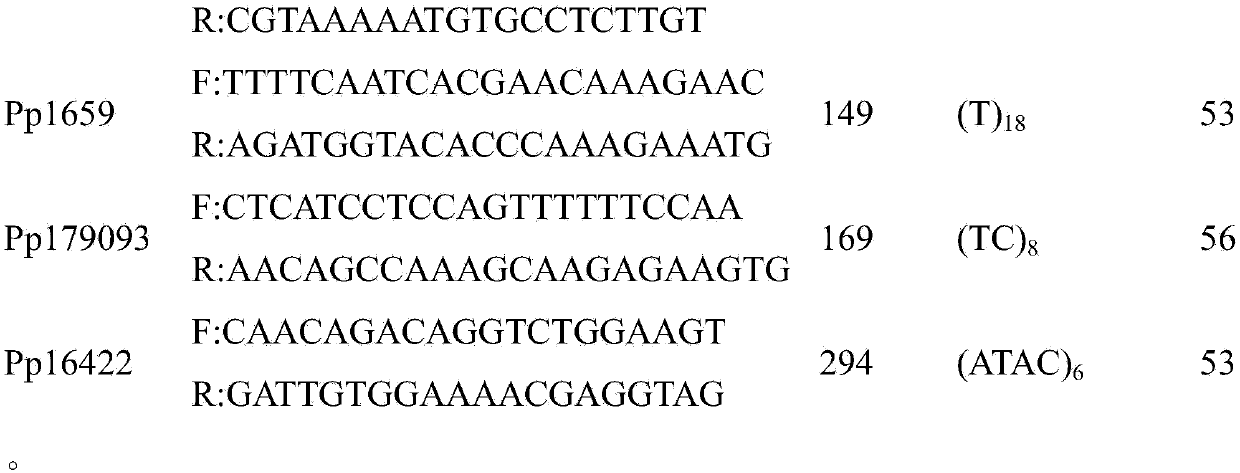Portunus pelagicus polymorphic microsatellite molecular marker, identification method and application
A technology of Portunus pelagics and microsatellite markers, which is applied to the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve problems such as high cost, low efficiency, and unsuitability for non-model animals, and achieve The effects of efficient identification, simple operation, and potential for widespread application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0025] 1. Extraction of Genomic DNA of Portunus pelagics
[0026] Collect 33 adult Portunus pelagics, take about 10g of muscle tissue and preserve it with 95% alcohol. Take about 100mg of muscle tissue, put it into a 1.5mL centrifuge tube filled with 300μL tissue lysate, and homogenate; add 10μL RNase (20mg / mL) to the centrifuge tube in turn, mix well, and incubate at room temperature for 2min; add 5μl proteinase K (20mg / mL), fully mixed, digested in a 55°C water bath until clarified, which can be shaken and mixed several times to accelerate its dissolution; followed by continuous extraction twice with Tris-saturated phenol and chloroform; absorb the supernatant Add about 300-350μL to a new centrifuge tube, add 1ml of pre-cooled absolute ethanol to precipitate DNA, centrifuge at 12000rpm to collect the precipitate, and wash it again with pre-cooled 70% ethanol; finally, dry the DNA (precipitation) at room temperature and dissolve in 40 μL sterile double-distilled water, and s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com