Application of oxathiapiprolin resistant gene RORP1 as selection marker during oomycetes conversion
A fluthiazolidin resistance and fluthiazolidin technology, which is applied in the field of genetic engineering, can solve the problems of limited screening markers, difficulty in re-knockout, and low screening efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Example 1. Application of Screening Marker RORP1 in Gene Complementation of Phytophthora capsici
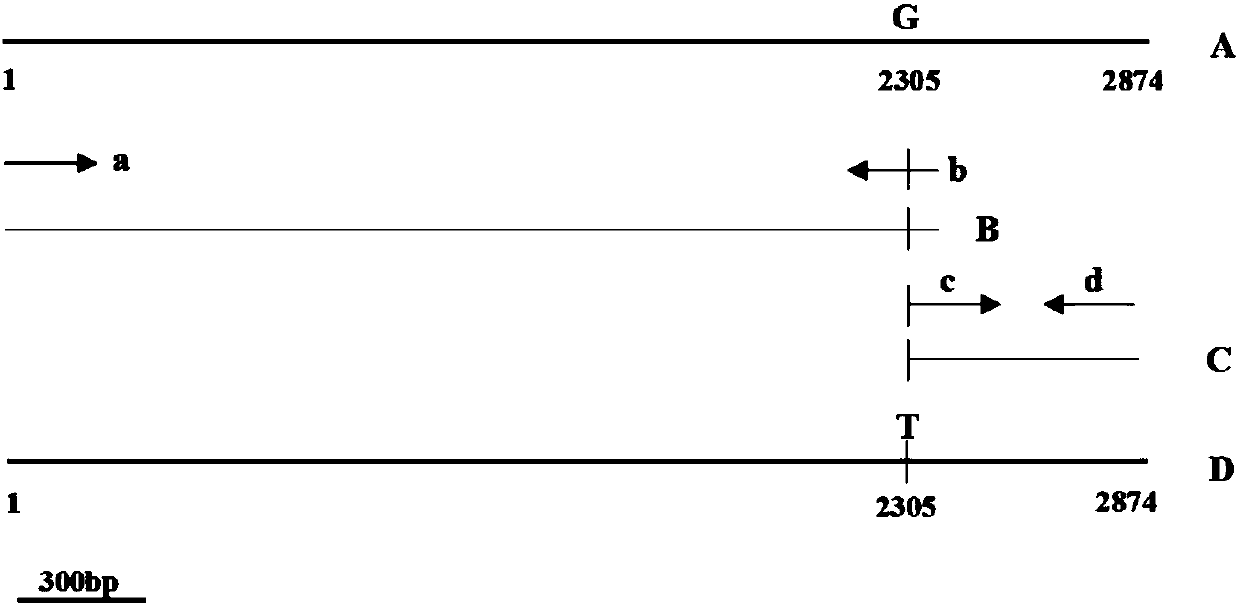
[0068] Since the sgRNA expressed by the sgRNA expression vector (pYF2.3G-N) used in the transformation of the above plasmid can target the NPTII gene, the vector can be used for complementation of the PcDHCR7 gene knockout transformant of Phytophthora capsici.
[0069] Construct the homology arm vector required for PcDHCR7 gene complementation; simultaneously transform the Cas9 protein expression vector pYF2-PsNLS-hSpCas9 (published vector, Fang and Tyler, 2016, Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR / Cas9 ), using the restriction endonuclease EcoR I to digest the pYF2-PsNLS-hSpCas9 vector, remove the NPT II gene and its promoter and terminator, and then connect with T4 ligase to obtain a Cas9 protein expression vector without the NPT II gene, Named pYF2-Cas9EI. The anteroporative homology arm vector and the...
Embodiment 2
[0077] Example 2, Application of Screening Marker RORP1 in Genetic Transformation of Phytophthora soybean
[0078] In order to expand the scope of application of the screening marker, the above-mentioned plasmid (pYF2.3G-RORP1-N) with the fluthiazolidone resistance gene was transferred into Phytophthora sojae protoplasts to verify whether the screening marker of the present invention can The method is used for the screening of transformants when it is applied to the genetic transformation of Phytophthora sojae, and the work efficiency when the screening marker is used for the genetic transformation of Phytophthora soybean.
[0079] The specific operation process is as follows:
[0080] The wild-type strain of Phytophthora sojae was used as the experimental material, and protoplasts were obtained by referring to the method of Fang et al., and the vector (pYF2.3G-RORP1-N) carrying the fluthiazolidin resistance gene was transferred into the protoplasts.
[0081] The protoplasts ...
Embodiment 3
[0085] Embodiment 3, the application of screening marker RORP1 in the genetic transformation of litchi peronosophthora
[0086] In order to further confirm the scope of application of the screening marker, the above-mentioned plasmid (pYF2.3G-RORP1-N) with the fluthiazolidin resistance gene was transferred into the protoplast of Lychee peronospora, to verify the screening marker of the present invention Whether it can be applied to the screening of transformants during genetic transformation of Pythophthora litchie, and the working efficiency of the screening marker when used for genetic transformation of Pythophthora litchie.
[0087] The specific operation process is as follows:
[0088] The wild-type strain of Phytophthora lychee was used as the experimental material, and protoplasts were obtained by referring to the method of Fang et al., and the vector (pYF2.3G-RORP1-N) carrying the fluthiazolidin resistance gene was transferred into the protoplasts.
[0089] The protopl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com