Voltage-gating sodium ion channel structure modeling method based on development coupling analysis
A sodium ion channel and coupling analysis technology, which is applied in the analysis of two-dimensional or three-dimensional molecular structure, electrical digital data processing, special data processing applications, etc., can solve the unresolved problems of three-dimensional structure, and overcome the poor prediction accuracy, High reliability, method reliable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
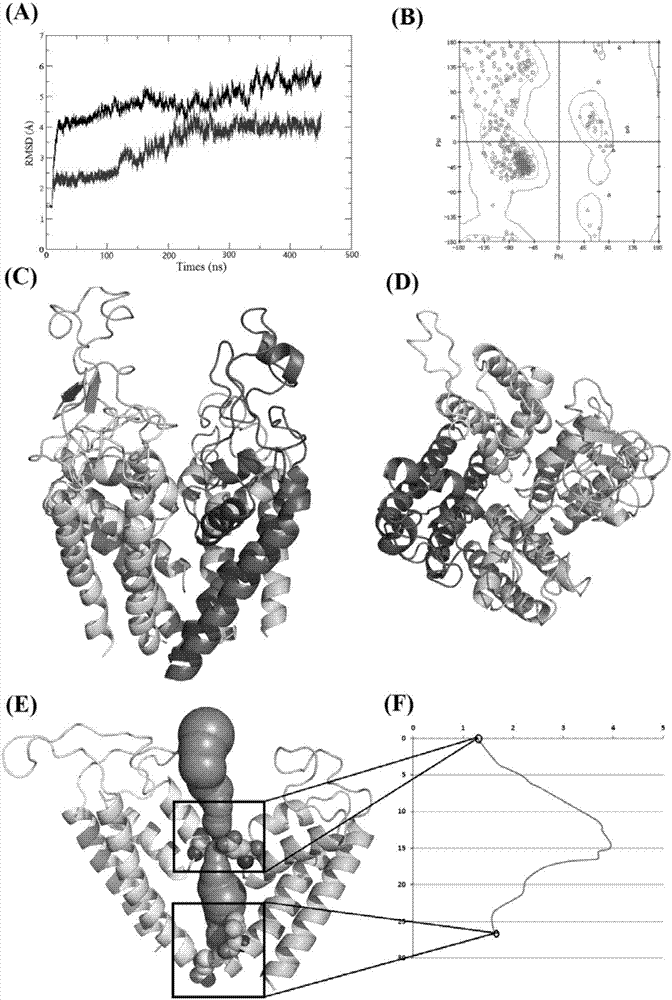
[0025] Embodiment 1: The structure modeling method for voltage-gated sodium ion channel of the present invention comprises the following steps:
[0026] 1) Construct four domain monomers and template structure Na respectively v Sequence alignment files between corresponding strands of PaS, such as figure 1 As shown, and using rosetta's membrane protein homology modeling method, the structure of the four domain monomers was constructed;
[0027] 2) By evolutionary coupling analysis, the interaction pairs present in the four monomers are predicted. The number of sequences that finally entered the multiple sequence alignment: DI: 1936, DII: 3923, DIII: 3390, DIV: 3597. The direct coupling analysis (DCA) value between residues in each monomer was calculated, and all residue pairs with a DCA value greater than 0.2 were retained as the final predicted interaction pair (CA_DCA).
[0028] 3) Calculate the interaction pairs within the four modeled domain monomer structures; If the ...
Embodiment 2
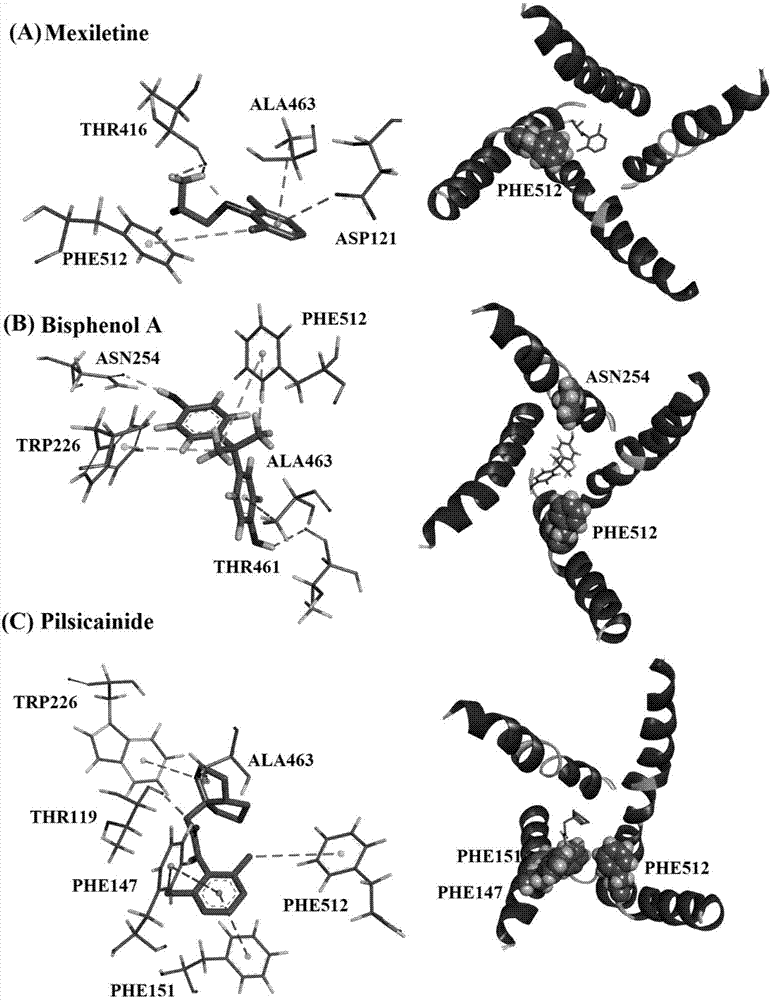
[0048] Example 2Na v 1.5 The interaction site with off-state local anesthetic drugs is consistent with the experimental results
[0049] 1)Na v 1.5 Complexes with off-state local anesthetics (Na v 1.5_CLA) construction
[0050] Choose to be able to Na v 1.5 Three local anesthetic molecules (CLA) acting in the off state: Pilsicainide, Bisphenol A and mexiletine as inhibitor molecules. The molecular docking method used is: use REDUCE, Autodock Tools and Autodoc4 to complete. Using CLA and Na v 1.5 A fully flexible docking method for the binding site region, using Autodock4 for docking. during the docking process. Scoring adopts the method of combining Autodock4's own scoring and Xscore scoring. Using the above method, three independent dockings were carried out for each drug molecule, and finally 57, 63 and 110 complex conformations were generated for the molecules of picicanilide, polyphenol A and mecillin, respectively. Select the molecular conformation with the lowes...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com