Efficient and stable bacterium total RNA (Ribonucleic Acid) extraction method
An extraction method, a bacterial technology, is applied in the field of microbial total RNA extraction, which can solve the problems of time-consuming, large differences, and poor extraction effects.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
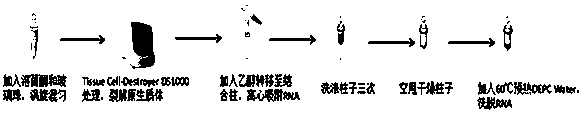
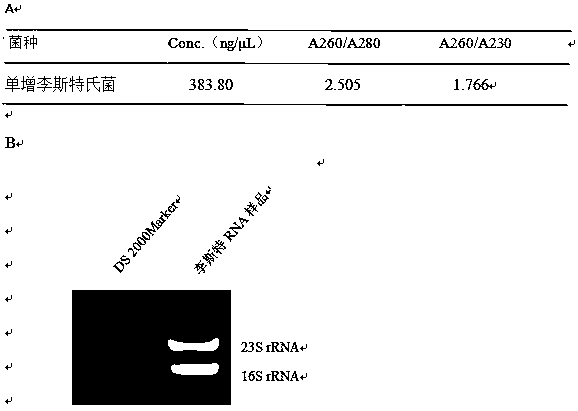
[0031] Embodiment 1: the extraction of Listeria monocytogenes RNA, described method specifically comprises the following steps:
[0032] The first step is as follows: prepare sterile TSB liquid medium, add Listeria monocytogenes with 1% inoculum, take out the bacterial liquid after shaking the bacteria at 37°C for 8 hours; place at 4°C for 10min; centrifuge at 5000×g After 10 minutes, the supernatant was discarded.
[0033] The second step is as follows: 200μL 15mg / mL lysozyme, 1mL blue tip to absorb appropriate glass bead powder into the extraction tube, vortex to mix, Tissue Cell-Destroyer DS1000 treatment, and its parameters are: Execute the project A, speed 5000RPM; time 30s; interval 10s; times 15.
[0034]The step three is specifically as follows: the kit used for subsequent processing is Bacterial RNA Kit R6950 (OMEGA), and 350 μL Buffer BRK / β-Me was added to the crushed solution, and vigorously vortexed for 5 minutes. Centrifuge at 13,000×g for 5 min; transfer 400 μL...
Embodiment 2
[0035] Embodiment 2: the extraction of Staphylococcus aureus RNA, described method specifically comprises the following steps:
[0036] The first step is as follows: prepare sterile TSB liquid medium, add Staphylococcus aureus to it with 3% inoculum, shake the bacteria at 37°C for 2h20min, then take out the bacterial liquid; place at 4°C for 10min; centrifuge at 5000×g for 10min, Aspirate and discard supernatant.
[0037] The second step is as follows: 200μL 15mg / mL lysozyme, 1mL blue shampoo, absorb appropriate glass bead powder into the extraction tube, vortex to mix, Tissue Cell-Destroyer DS1000 treatment, and its parameters are: Execute the project A, speed 5000RPM; time 30s; interval 10s; times 15.
[0038] The step three is specifically as follows: the kit used for subsequent processing is Bacterial RNA Kit R6950 (OMEGA), and 350 μL Buffer BRK / β-Me was added to the crushed solution, and vigorously vortexed for 5 minutes. Centrifuge at 13,000×g for 5 min; transfer 400 μ...
Embodiment 3
[0039] Embodiment 3: the extraction of Klebsiella pneumoniae RNA in the blood sample, described method specifically comprises the following steps:
[0040] The step 1 is specifically as follows: draw 2 mL of blood sample into a 15 mL centrifuge tube with a 5 mL disposable syringe, add ACKLysing Buffer, ice-bath for 30 min, and mix by inverting up and down every 5 min. 12000rpm / min, centrifuge for 10min, discard the supernatant. Resuspend the pellet in TE buffer and vortex to mix. Centrifuge at 5000×g for 10 minutes, discard the supernatant.
[0041] The second step is as follows: 300μL 15mg / mL lysozyme, 1mL blue tip to absorb appropriate glass bead powder into the extraction tube, vortex to mix, Tissue Cell-Destroyer DS1000 treatment, and its parameters are: Execute the project A, speed 5000RPM; time 30s; interval 10s; times 15.
[0042] The step three is specifically as follows: the kit used for subsequent processing is Bacterial RNA Kit R6950 (OMEGA), and 350 μL Buffer BR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com