Probe and kit for detecting SNP (Single Nucleotide Polymorphism) site by utilizing recombinase mediated isothermal amplification method based on probe guidance
A technology of isothermal amplification and recombinant enzymes, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problems with high requirements and achieve rapid results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
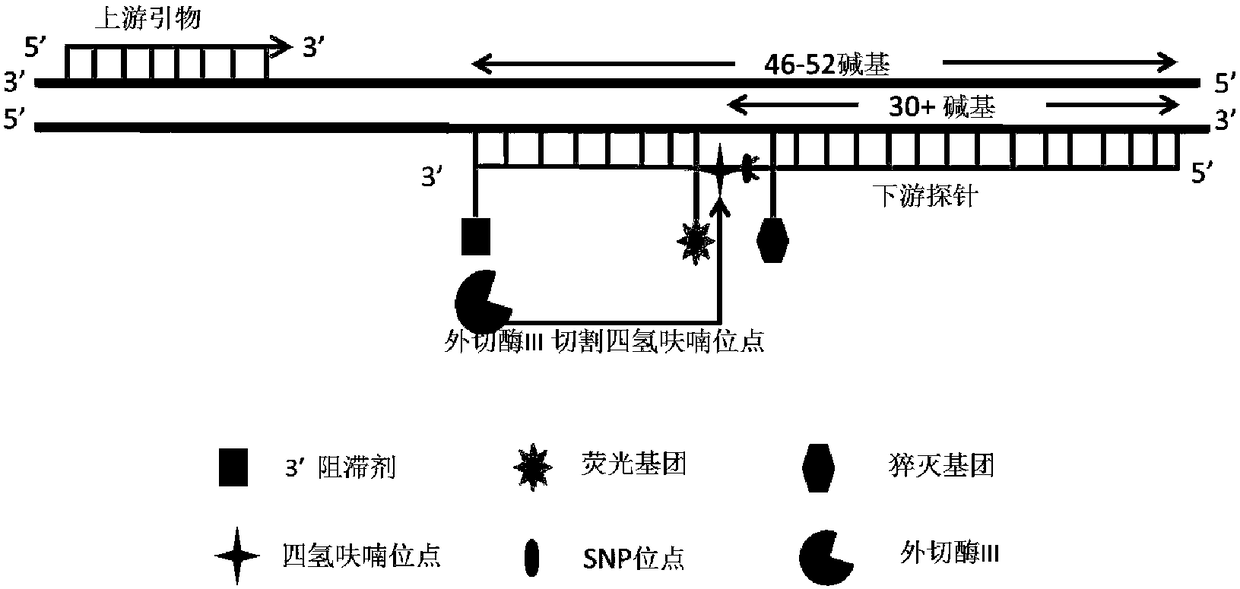
[0048] The flanking sequence of the MTHFRA1298C SNP site was obtained through the dbSNP database of the National Center for Biotechnology Information (NCBI), and a primer and two primers suitable for this method were designed according to the design principles of the RAA method using Oligo7.37 software. probe. At the same time, in addition to the design principles of RAA, we require that the SNP site should be located just before the THF site of the probe, such as figure 1 shown. All primers and probes were synthesized by Shanghai Sangon Bioengineering Co., Ltd. and purified by high performance liquid chromatography. PRAA primers and probes are listed in Table 1.
[0049] Table 1 PRAA primers and probes
[0050]
[0051] a. Forward primers used in A and C reactions
[0052] b. Specific probes for A reactions
[0053] c. Specific probes for C reactions
[0054] d. Modification of the probe: FAM, 6-Carboxyfluorescein (6-carboxyfluorescein); THF, tetrahydrofuran (tetrahy...
Embodiment 2
[0056] 1. Collection of clinical specimens
[0057] From 2016 to 2017, 150 children with CHD hospitalized in Children's Cardiac Surgery in Hebei Province were collected, including 72 males and 78 females, aged 1 month to 14 years. The whole blood DNA of the specimen was extracted with the Tiangen Biochemical Blood Genomic DNA Extraction Kit produced by Tiangen Biochemical Technology (Beijing) Co., Ltd., the genomic DNA was dissolved in 150 μL of nuclease-free water, and stored at -20°C until use. The DNA concentration of the obtained samples ranged from 10–75 ng / μL. This study was approved by the Medical Ethics Committee of Hebei Children's Hospital, and the parents of the children signed the informed consent.
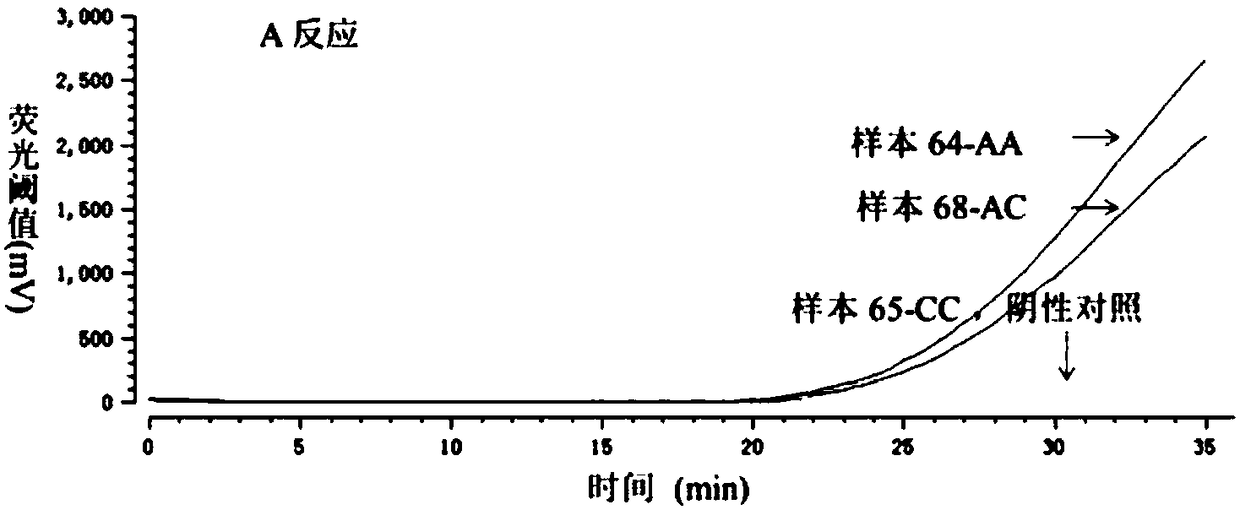
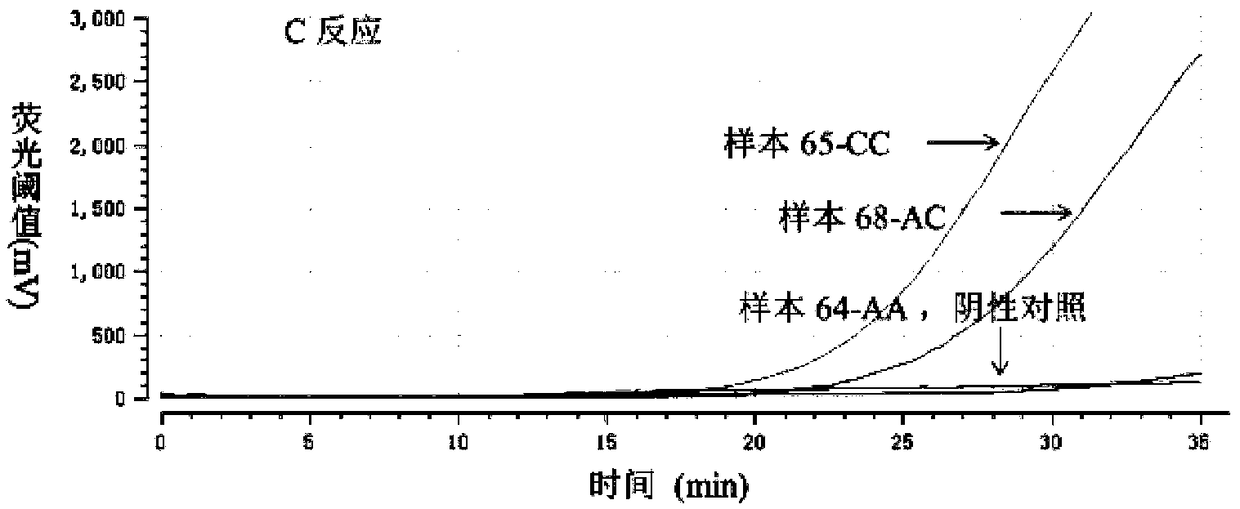
[0058] 2. Perform PRAA detection on 150 clinical samples
[0059] The RAAExo kit produced by Jiangsu Qitian Gene Company was used. Both A and C reactions were carried out in a 0.2ml reaction tube, which included a pre-lyophilized enzyme mixture (SSB, UvsX, DNA polym...
Embodiment 3
[0068] In order to further explore the effect of sample concentration on the specificity of the PRAA method, we constructed two plasmids carrying homozygous AA and CC homozygotes at this site, and made a series of dilutions, including 10 7 ,10 6 ,10 5 ,5*10 4 ,10 4 ,5*10 3 ,10 3 ,5*10 2 ,10 2 The copies / reactions were then tested separately using the PRAA method (A and C reactions).
[0069] Use the AA type plasmid to explore the sensitivity of the A reaction and the specificity of the C reaction, and use the CC type plasmid to explore the specificity of the A reaction and the sensitivity of the C reaction. For plasmids, we found that the lower limit of templates that can be detected by reactions A and C are both 5*10 3 copy each reaction, ( Figure 3-1 with Figure 3-1 ). Non-specific amplification will occur when the amount of mismatched template reaches 105 copies per reaction ( Figure 3-3 with Figure 3-4 ). It is suggested that the PRAA method may be locate...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com