Double digital PCR (polymerase chain reaction) method of African struthio camelus derived ingredient quantitative detection
A technology for quantitative detection of source components, applied in the field of molecular biology detection, can solve problems such as no quantitative detection methods, save reagents and time costs, and avoid systematic errors.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0065] 1. Preparation of samples and extraction of DNA templates: Take 25-30 g of samples (meat or meat-based foods, etc.) and cut them into pieces, then use a tissue grinder to crush them. The crushing conditions are 1800 rpm for 3 minutes. Weigh 20mg~50mg of the prepared sample into a 1.5mL centrifuge tube, and extract the DNA of the sample using the kit method. Promega, A1120), PSS nucleic acid automatic extractor and other DNA extraction methods.
[0066] 2. Preparation and dispersion of reaction system
[0067] (1) The reaction system of ddPCR digital PCR is:
[0068] The ddPCR (Droplet digital PCR) reaction system is 20 μL, and the components are as follows: 2×ddPCR TM 10 μL of master mix; 0.8 μL of each primer with a concentration of 10 μmol / μL, 0.4 μL of each probe with a concentration of 10 μmol / μL, 2 μL of DNA template, and replenish water to 20 μL.
[0069] Add 20 μL of the reaction system and 70 μL of droplet generation oil into the droplet generation card slot,...
Embodiment 1
[0103] Example 1 Verification of the Relative Qualitative Detection Limit of the African Ostrich-derived Component Copy Number Concentration
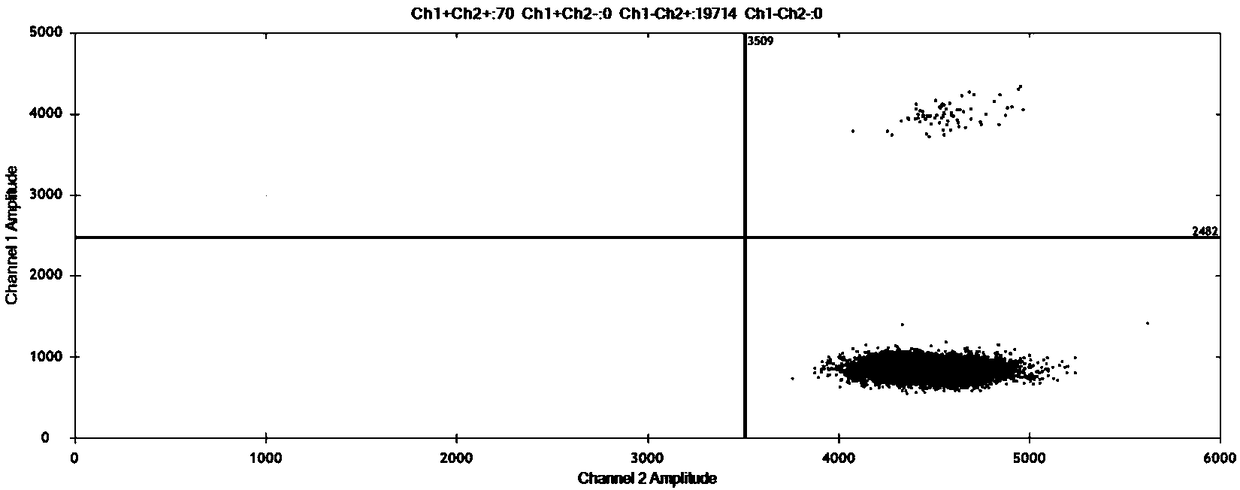
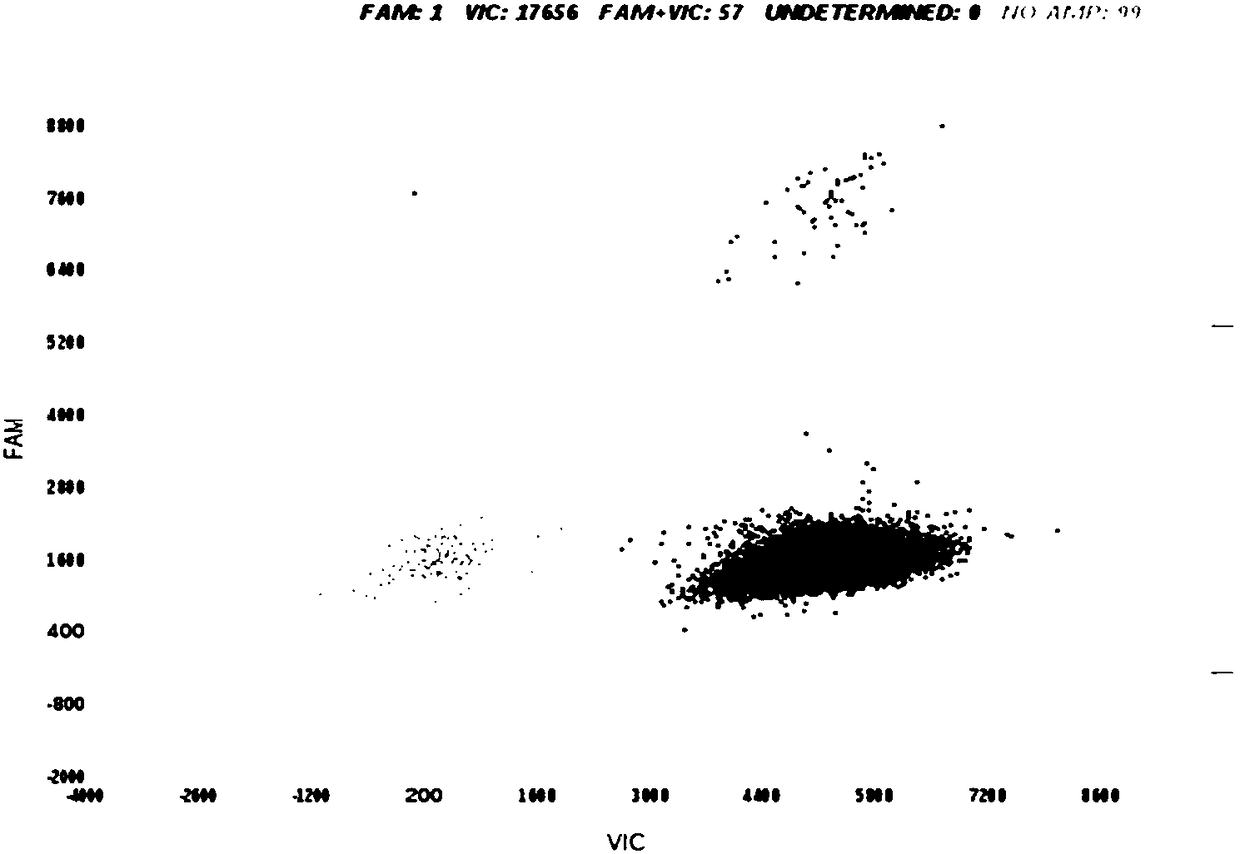
[0104] Samples to be tested: Genomic DNA from pigs, cattle, sheep, and chickens is used as a matrix, and African ostrich genomic DNA is mixed according to the copy number percentage to form a test DNA sample with a copy number percentage of African ostrich genomic DNA of 0.01%. Three parallel ddPCR and cdPCR experiments were carried out, and the obtained data are shown in figure 1 and figure 2 . The results showed that both ddPCR and cdPCR could be detected when the content of African ostrich-derived components was 0.01%.
Embodiment 2
[0105] Example 2 Verification of the Relative Quantitative Detection Limit of the Copy Number Concentration of African Ostrich-derived Components
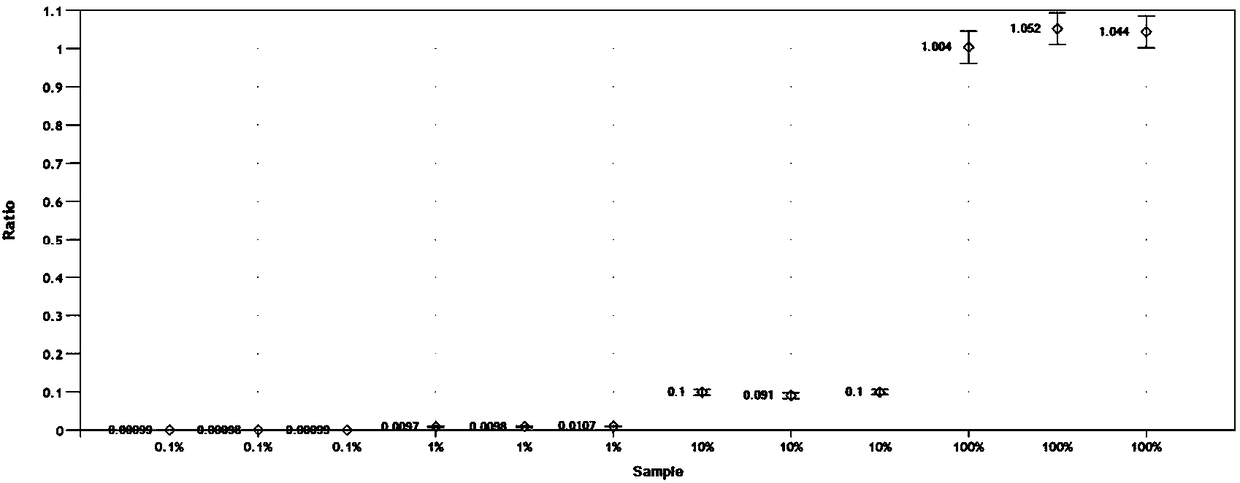
[0106] Samples to be tested: In order to verify the quantitative detection limit of this method, pig, cow, sheep, and chicken genomic DNA were used as substrates, and African ostrich genomes with copy number percentages of 0.1%, 1%, 10%, and 100% were incorporated into them DNA. Three parallel ddPCR and cdPCR experiments were carried out respectively, and the obtained experimental results are shown in image 3 and Figure 4 .
[0107] For African ostrich genomic DNA samples with copy number percentages of 0.1%, 1%, 10% and 100%, the detection results on the ddPCR platform were 0.098%, 1.062%, 9.697% and 101.34%, respectively, and the three parallel The RSD value was between 1.03% and 5.34%, and the recovery rate was between 96.97% and 101.34%. The RSD value is between 3.08% and 14.11%, and the recovery rate is between 97.91% an...
PUM
| Property | Measurement | Unit |
|---|---|---|
| recovery rate | aaaaa | aaaaa |
| recovery rate | aaaaa | aaaaa |
| recovery rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com