Renal clear cell carcinoma related biomarkers and their application
A clear cell renal cell carcinoma and cell technology, which is applied in the field of biomarkers, can solve the problems that molecular diagnosis and targeted therapy of renal clear cell carcinoma cannot be satisfied, and the prognosis is not very ideal.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
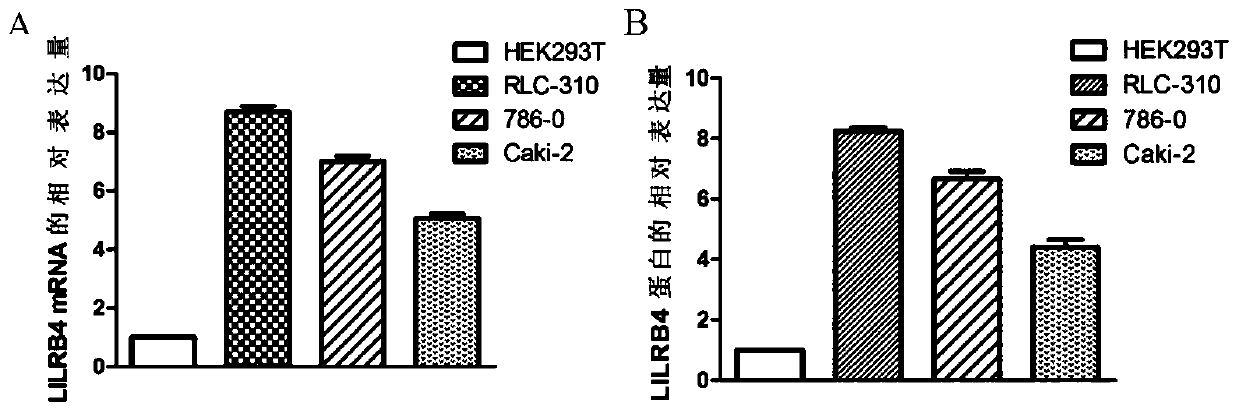
[0087] Example 1 Screening for Gene Markers Related to Renal Clear Cell Carcinoma
[0088] 1. Sample collection
[0089]Tissues from 6 cases of clear cell renal cell carcinoma and corresponding paracancerous tissues, and renal tissues from 4 patients with non-clear cell renal cell carcinoma were collected. All patients had not received radiotherapy and chemotherapy before operation, and postoperative pathological examination was performed to confirm the diagnosis. The informed consent of the patients was obtained for the acquisition of tissue samples, and the consent of the organizational ethics committee was obtained.
[0090] 2. Preparation of RNA samples
[0091] RNA samples were extracted using the QIAGEN Tissue RNA Extraction Kit. For details, please refer to the instruction manual.
[0092] 3. Remove rRNA
[0093] Ribosomal RNA was removed from total RNA using the Ribo-Zero kit.
[0094] 4. Construction of cDNA library
[0095] The cDNA library was constructed using...
Embodiment 2
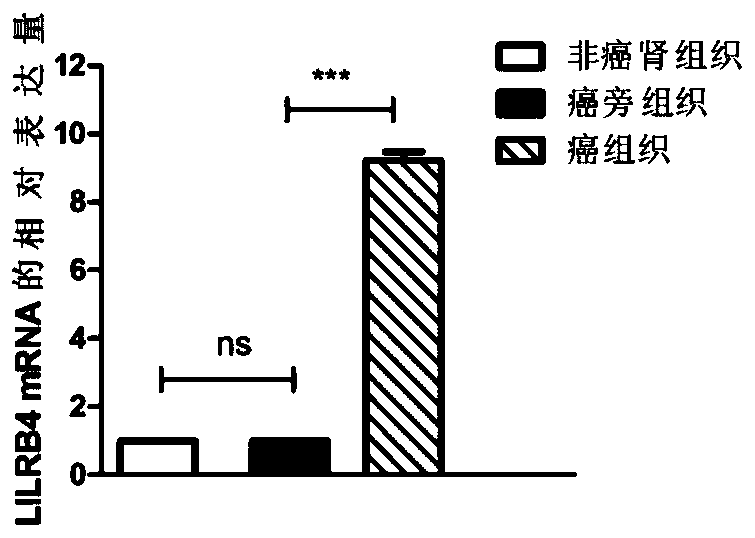
[0102] Example 2 QPCR sequencing to verify the differential expression of the LILRB4 gene
[0103] 1. Large-sample QPCR verification of differential expression of LILRB4 gene. According to the sample collection method in Example 1, 50 cases of paracancerous tissues and clear cell carcinoma tissues of renal clear cell carcinoma patients were selected, and 18 cases of renal tissues of non-renal clear cell carcinoma patients were collected.
[0104] 2. RNA extraction
[0105] RNA samples were extracted using the QIAGEN Tissue RNA Extraction Kit. For details, please refer to the instruction manual.
[0106] 3. Reverse transcription
[0107] mRNA reverse transcription was performed using the FastQuant cDNA First Strand Synthesis Kit (Product No.: KR106). Specific steps are as follows:
[0108] (1) Add 5×gDNA Buffer 2.0μl, total RNA 1μg, add RNase Free ddH 2 O to bring the total volume to 10 μl, heat in a water bath at 42°C for 3 minutes;
[0109] (2) Construct a 20μl reaction...
Embodiment 3
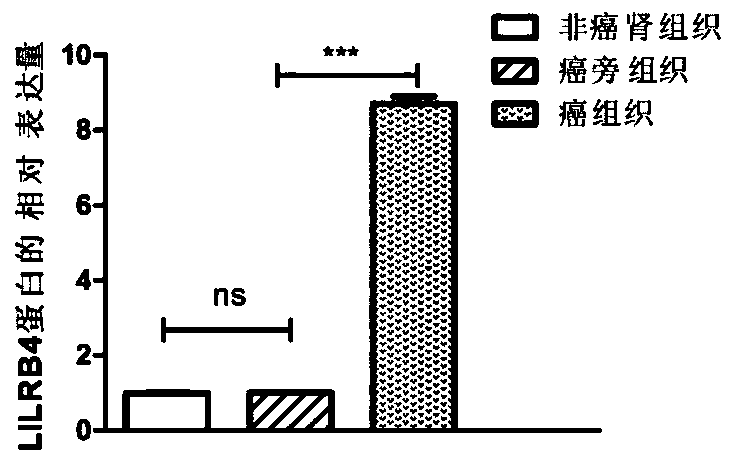
[0124] Example 3 Western blotting assay to detect the differential expression of LILRB4 protein
[0125] 1. Extraction of total tissue protein
[0126] Cut the tissue with scissors and put it into a glass homogenizer placed in ice. Mix the RIPA lysate and PMSF at a ratio of 100:1, and add the corresponding amount of RIPA lysate, crush the tissue with a glass homogenizer until it is fully lysed, suck the lysed liquid into an EP tube, centrifuge at 14,000 rpm for 5 min at 4°C, and collect the supernatant.
[0127] 2. Determination of total protein concentration
[0128] The protein concentration was determined according to the instructions of the BCA protein concentration determination kit.
[0129] 3. SDS-PAGE electrophoresis
[0130] Prepare 8% separating gel and 5% stacking gel according to the instructions of the SDS-PAGE gel preparation kit and perform electrophoresis.
[0131] 4. Western detection
[0132] 1) Electrotransfer
[0133] Put the PVDF membrane into methan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com