DNA sequencing method using nucleotide and nucleotide with reversibly blocked end 3'
A DNA sequencing and nucleotide technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve problems such as unknown, achieve the effects of improving accuracy, improving sequencing accuracy, and reducing sequencing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
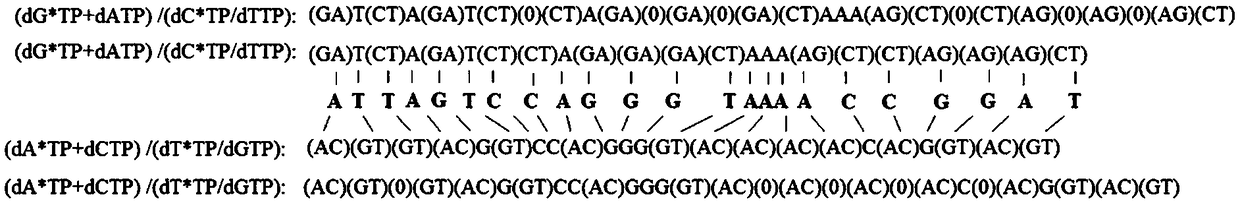
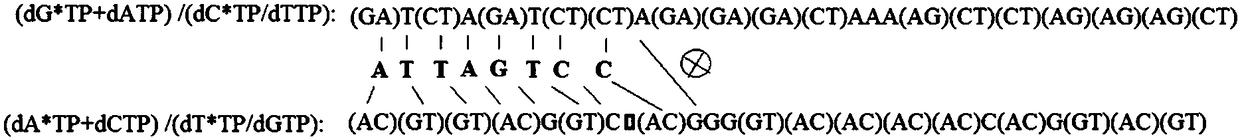
[0032] Example 1: A DNA sequencing method comprising nucleotides and 3' end reversibly blocked nucleotides was used to determine the artificially synthesized sequence comprising 3'-TAATCAG GTCTG-5' fragments.
[0033] 1. Template preparation: the artificially synthesized templates modified with 5' biotin were immobilized with avidin-modified magnetic beads, and then the magnetic beads were separated from the liquid, and the artificially synthesized templates immobilized on the magnetic beads were used for hybridization with sequencing primers.
[0034] 2. Hybridization of sequencing primers: Incubate the designed sequencing primers with the template immobilized by magnetic beads at 75°C for 5 minutes, then cool naturally to room temperature, then separate the magnetic beads from the liquid, and use the template immobilized by magnetic beads for DNA sequencing.
[0035] 3. Place the template immobilized by magnetic beads in the reactor (block both ends of the reactor with a semi...
Embodiment 2
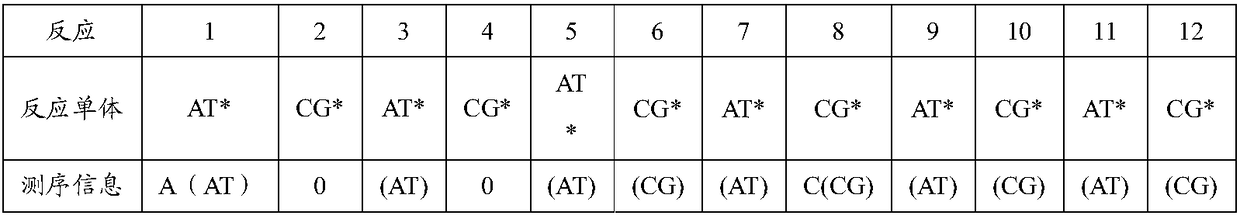
[0058] Example 2: Two-nucleotide real-time synthetic DNA decoding and sequencing of the Escherichia coli genome
[0059] 1. Preparation of the whole genome template: the Escherichia coli genome is broken into fragments with a size of 100-1000 bp bases by ultrasound, and these fragmented nucleic acid sequences are connected with a pair of universal linkers with known sequences (such as : The sequence of the linker 1 is: CTG CTG TAC CGT ACA GCC TTG GCC G; the sequence of the linker 2 is: CGC TTT CCTCTC TAT GGG CAG TCG GTGA T) for ligation and pre-amplification for 10 cycles; then gel The 200-800bp DNA fragment was cut by electrophoresis and purified. These 200-800bp DNA fragments were subjected to emulsion parallel PCR reaction with microbeads immobilized with complementary sequences of one of the linkers to amplify the fragmented Escherichia coli genome fragments and denature them to obtain Escherichia coli genome sequencing DNA templates Beads of double-stranded DNA templates...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com