Kit for detecting X-STR gene seat based on next generation sequencing technology and dedicated primer combination thereof
A technology of X-STR and primer combination, applied in the field of forensic medicine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0125] Example 1. Preparation of a kit for detecting the X-STR locus based on next-generation sequencing technology
[0126] 1. Preparation of primer combinations
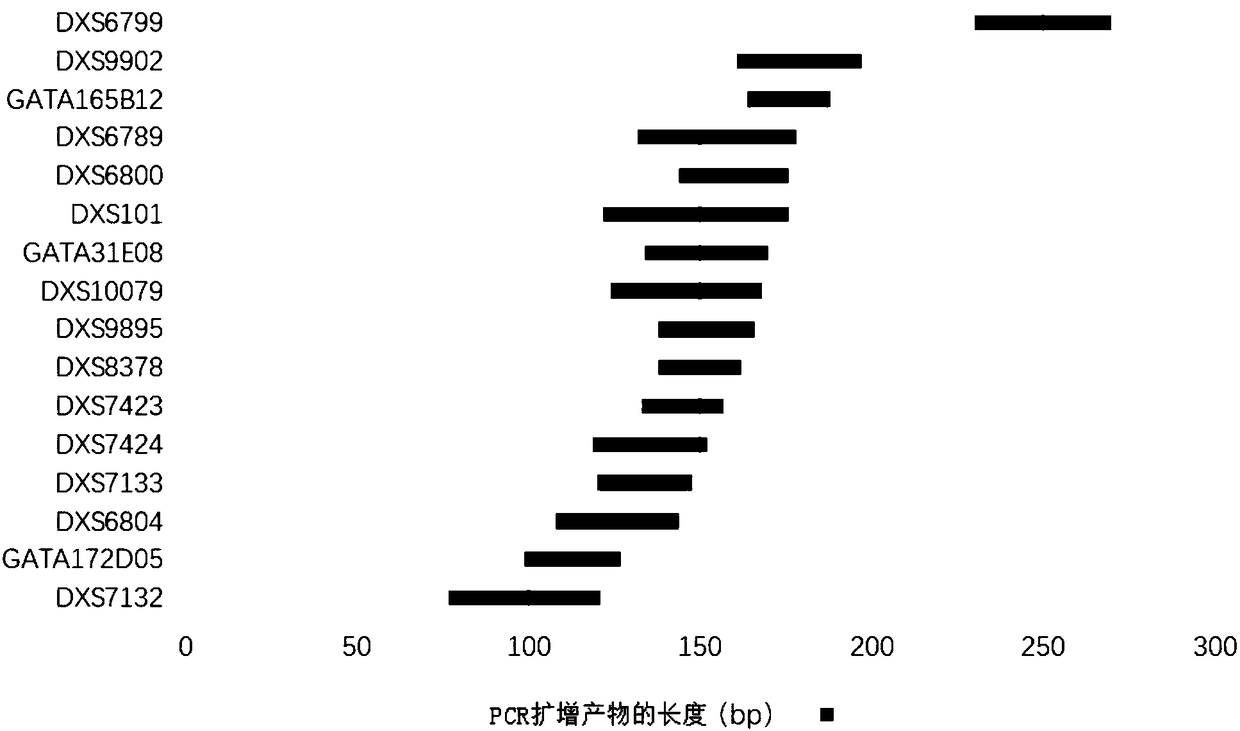
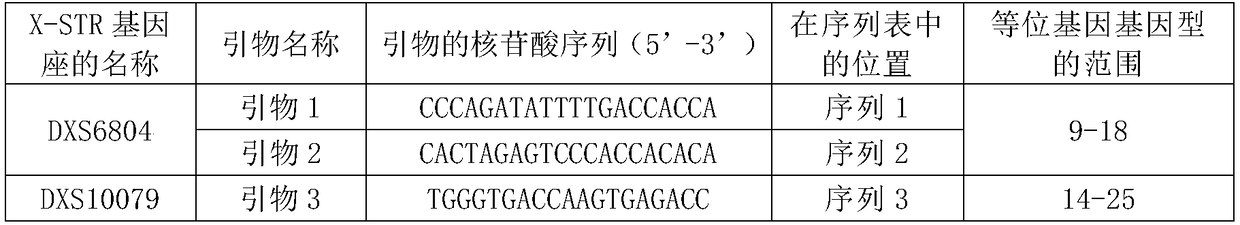
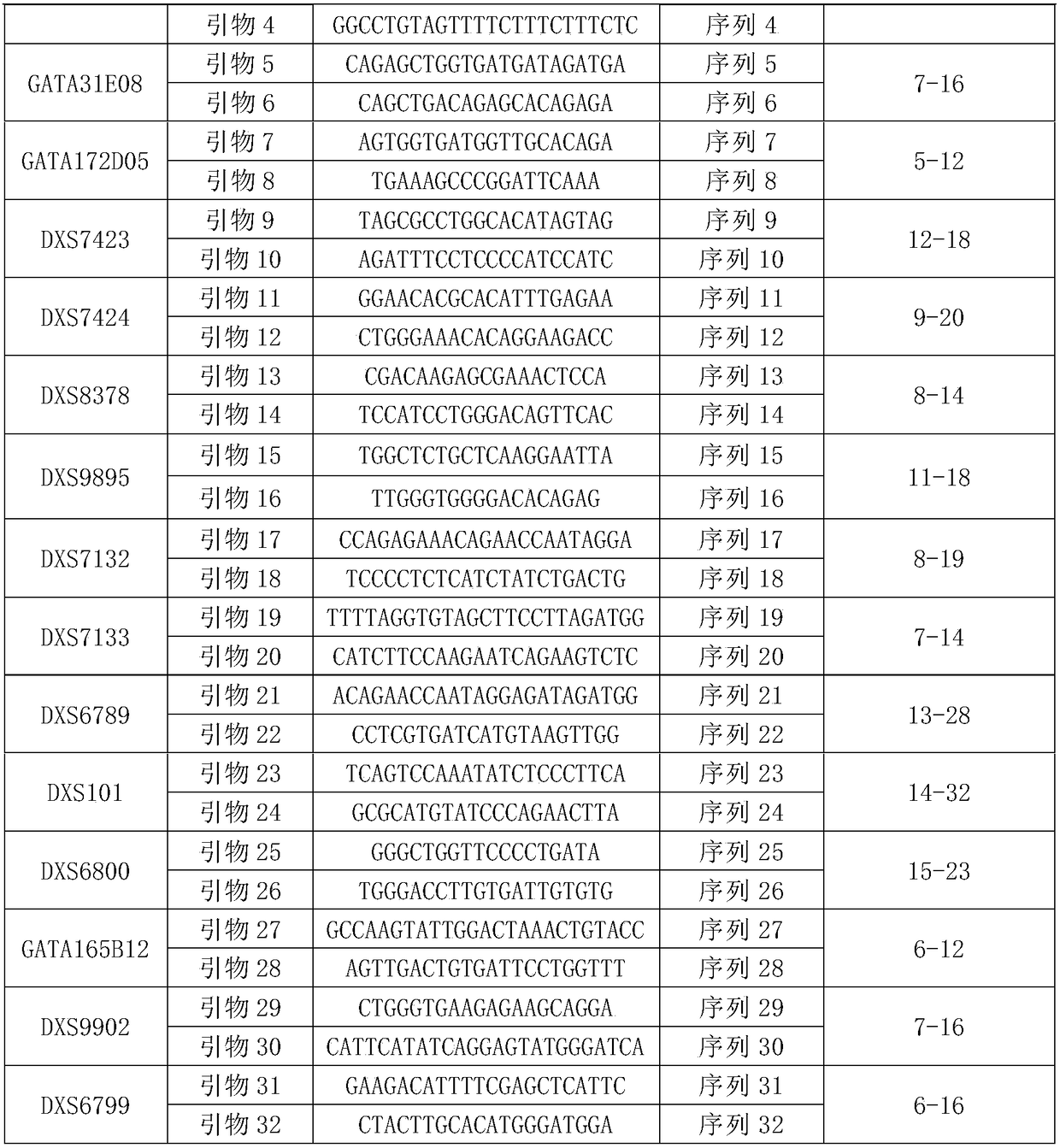
[0127] The primer set consisted of 32 primers for the detection of 16 X-STR loci. The name of each X-STR locus, the name of the primer corresponding to the amplified X-STR locus, the nucleotide sequence of the primer, and the range of allelic genotypes are shown in Table 1. The primers corresponding to each X-STR locus are the PCR amplification products in the hg19 human reference genome (for information on the hg19 human genome, see the website http: / / hgdownload.soe.ucsc.edu / goldenPath / hg19 / bigZips / hg19.2bit) See Table 2 for the length and nucleotide sequence.
[0128] Table 1
[0129]
[0130]
[0131] Table 2
[0132]
[0133]
[0134] 2. Preparation of kits for detecting X-STR loci based on next-generation sequencing technology
[0135] The kit for detecting the X-STR locus based on next-genera...
Embodiment 2
[0136] Example 2. Application of the kit for detecting the X-STR locus based on next-generation sequencing technology
[0137] 1. DNA sample preparation
[0138] Genomic DNA of a female oral epithelial cell sample was extracted, and then diluted with ultrapure water to obtain an aqueous solution of genomic DNA of female oral epithelial cells with a concentration of 0.425 ng / μL.
[0139] 2. Library preparation
[0140] 1. PCR amplification
[0141] Using the aqueous genomic DNA solution of female oral epithelial cells obtained in Step 1 as a template and the primer mixture prepared in Step 2 of Example 1 as primers, PCR amplification was performed to obtain PCR amplification products.
[0142] The reaction system is 20 μL, consisting of 10 μL Master Mix (product of the Physical Evidence Identification Center of the Ministry of Public Security), 4 μL primer mix, 2.5 μL genomic DNA aqueous solution of female oral epithelial cells and 3.5 μL ddH 2 O composition. In this reacti...
Embodiment 3
[0154] Example 3. Verification of the accuracy of the kit for detecting the X-STR locus based on next-generation sequencing technology
[0155] 1. DNA sample preparation
[0156] Genomic DNA of a female oral epithelial cell sample was extracted, and then diluted with ultrapure water to obtain an aqueous solution of genomic DNA of female oral epithelial cells with a concentration of 0.425 ng / μL.
[0157] Second, capillary electrophoresis detection
[0158] Take 1ng of the genomic DNA of female oral epithelial cells obtained in Step 1, and perform capillary electrophoresis detection according to the instructions of DNATyper X19 (product of the Physical Evidence Identification Center of the Ministry of Public Security), to obtain the allelic genotype of the locus. The typing results are detailed in column 2 of Table 4.
[0159] Third, next generation sequencing technology detection
[0160] The genomic DNA of the female oral epithelial cells obtained in step 1 was detected acc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com