A haplotype marker associated with rapid growth in largemouth bass and its application
A technology for largemouth bass and genotype, which is applied in the field of haplotype marking, can solve the problems of large growth disparity, mutual cannibalism, and insufficient parental quality of largemouth bass, and achieves fast growth rate, direct effect, and reduced blindness effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1 Obtaining SNP markers related to growth traits of largemouth bass
[0062] The present application found that the amino acid sequence of the largemouth bass HSP70 gene (heat shock protein 70) is shown in SEQ ID NO: 1; its cDNA sequence is shown in SEQ ID NO: 2, wherein the 993rd base is G or A; its gene sequence is shown in SEQ ID NO: 3, which consists of coding region and non-coding region, wherein the 1313th R is the base G or A, and there are two genotypes GA and AA; the 259th K is the base The base T is either a base deletion, and there are two genotypes T- and -- (where "-" indicates a base deletion); the 2302nd S is a base segment CTGTCTTCTGTTCATAAGTGTC (abbreviated as a Y segment) or a base segment deletion , that is, there are two genotypes Y- and -- (where "-" means a base deletion); the 2346th W is a base A or T, and there are two genotypes AT and AA.
[0063] The random population of 430 samples used for correlation analysis in this experiment was ...
Embodiment 2
[0074] Embodiment 2 A kind of method of screening fast-growing largemouth bass
[0075] Utilize above-mentioned SNP locus to screen fast-growing largemouth bass parent, comprise the following steps:
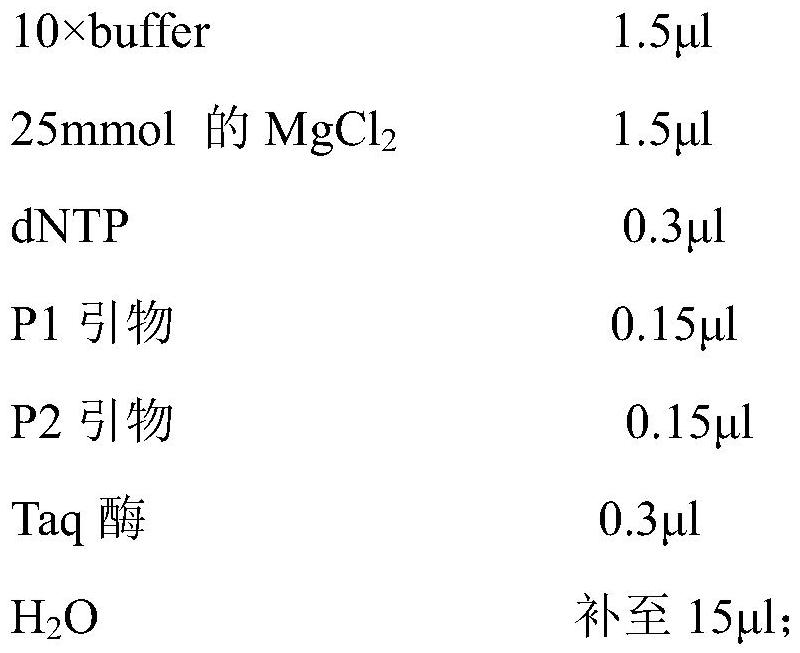
[0076] (1) Primer sequence:
[0077] According to the largemouth bass heat shock protein HSP70 gene sequence, a pair of primers were designed for PCR amplification. The designed and synthesized primers were as follows:
[0078] P1: 5'-CGTCGACTTCTACACCTCCA-3' (SEQ ID NO: 4)
[0079] P2: 5'-TCGTCTGGGTTGATGCTCTT-3' (SEQ ID NO:5)
[0080] The primers are expected to amplify a DNA band with a size of 231bp.
[0081] (2) Sample treatment (alkali lysis method):
[0082] 1. Cut 10-20 mg of fin rays to be tested and put them into a clean EP tube;
[0083] 2. Add 180μL of 50mmol / L NaOH solution, bathe in water for 20min (room temperature), shake several times during the period;
[0084] 3. Add 20 μL of 1mol / L Tris-HCL solution (PH=8.0), and vortex fully;
[0085] 4. Put the sample t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com