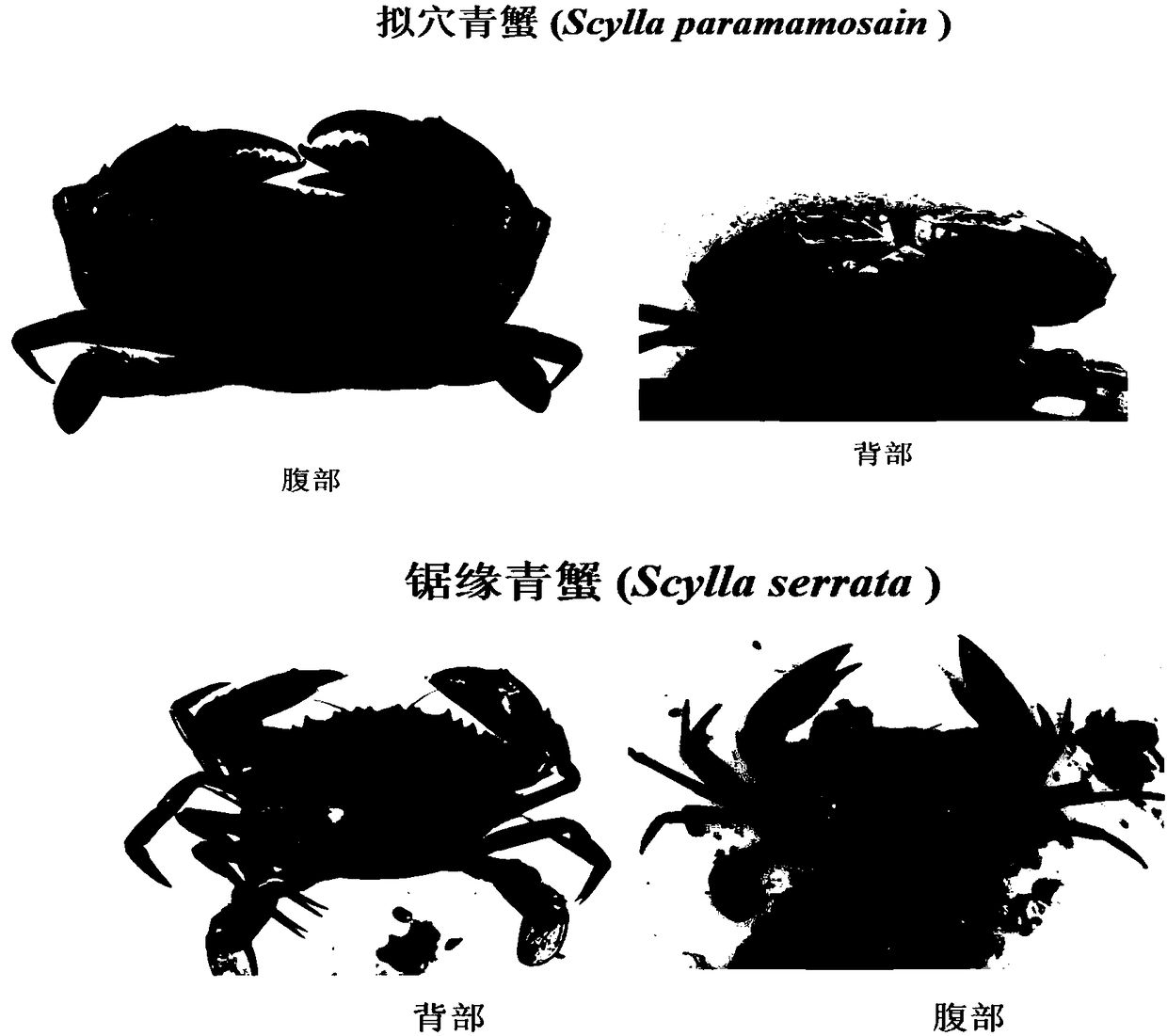
Method for identifying scylla paramamosain from scylla serrate
A technology of Scylla serrata and Scylla serrata, which is applied in the field of identifying Scylla serrata and Scylla serrata, can solve the problems of resource decline of wild blue crabs and imminent artificial breeding, etc., and achieve the effect of accurate identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Screening of Molecular Markers for Distinguishing Scylla pseudophylla and Scylla serrata
[0026] Obtain the mtDNA sequences of Scylla pseudocavena and Scylla serrata in the NCBI database, and use AlignX (a component of Vector NTI Suite 7.1) software to perform sequence alignment to find out the difference base region in Scylla; finally screen the obtained consensus Sequence, in the interval 13274-13362 of the consensus sequence, based on the consensus sequence, primers were designed, PCR was amplified and verified by sequencing, and the counted SNP position was 13334, a total of 1 SNP site. The sequence of the corresponding nucleotide fragment in Scylla paramamosain comprising the above SNP site is SEQ ID NO: 1:
[0027] AATTAAAAAACTAATGATTATGCTACCTTTGCACGGTCAAAATACCGCGGCTATTTAACA-TTTTTGTCAGTGAGCAGGCTAGACTATA;
[0028] The sequence of the corresponding nucleotide fragment in Scylla serrata (Scylla_serrata) is SEQ ID NO: 2:
[0029] AATTAAAAAACTAATGATTATGCT...
Embodiment 2
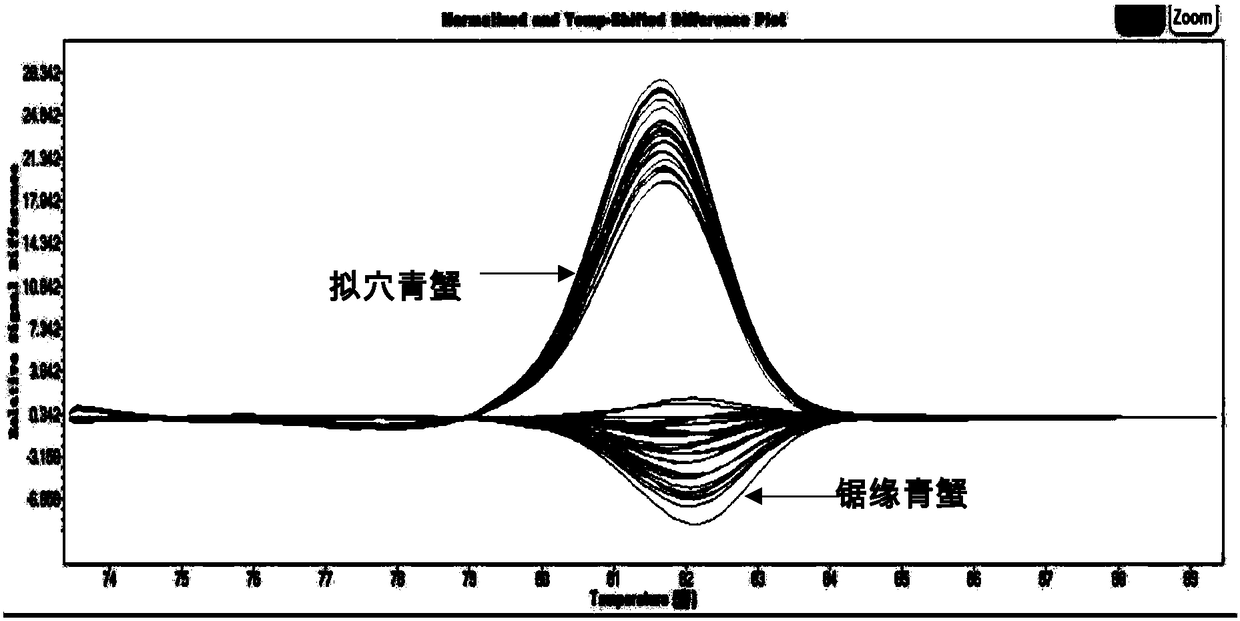
[0037]Example 2 Utilize SNP sites to identify blue crabs
[0038] The mud crabs used, mud crabs serrata, and mud crabs were collected from healthy individuals in Lulin Market, Ningbo City, China, and Sanmen County, China, respectively. The morphological identification was carried out according to the morphological characteristics of mud crabs studied by Lin Qi et al. Take 30 individuals of each variety, take swimming foot muscle tissue respectively, use phenol-chloroform method to extract genomic DNA, dilute the extracted DNA to 100ng / microliter, and use 1.5% agarose gel electrophoresis to detect its integrity, save Store at -20°C for later use.
[0039] PCR reactions were performed in a Roche Light Cycle 480 instrument. The reaction system is 20 microliters, including 10 microliters LC-HRM master Mix 10 microliters, forward and reverse primers 1.5 microliters, template DNA 1.2 microliters, grade H 2 O 3 µl. The reaction program was pre-denaturation at 95°C for 10 min, dena...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com