InDel molecular marker related to cashmere traits of goat and application of InDel molecular marker
A molecular marker, goat technology, applied in the field of molecular biology, to achieve the effect of accurate and reliable method, promoting the breeding process and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Genome-wide selection signal analysis of cashmere-producing goats and non-cashmere-producing goats
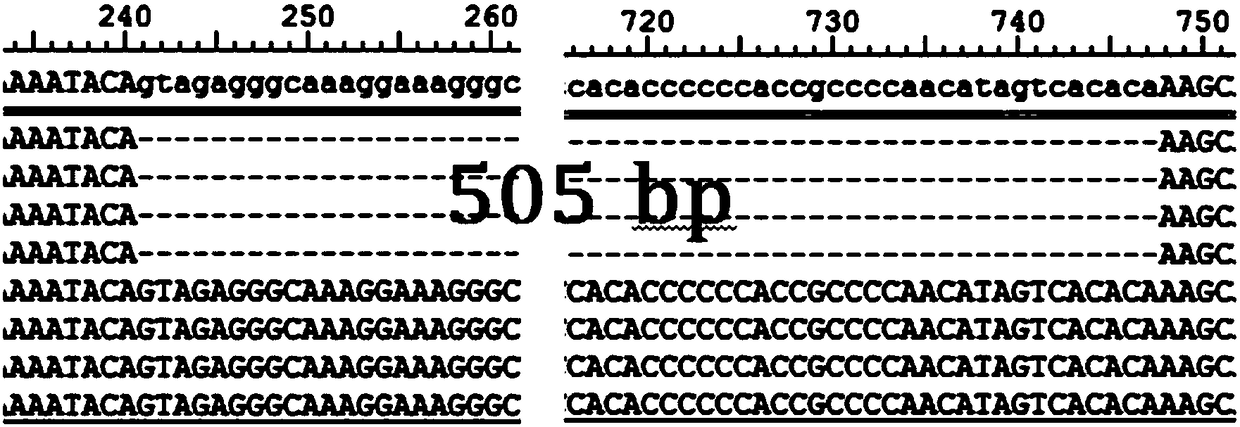
[0042] The whole-genome resequencing data of 66 non-cashmere goats in Central Pakistan, Nepal, and China and 51 cashmere-producing goats from China were analyzed for the gene-based selection signal of cashmere-non-cashmere production, and it was found that it is located on chromosome 6 of goats. The FGF5 gene locus was most closely related to cashmere-non-downy traits. Through further data mining, it was found that the InDel molecular marker located at 95454685-95455189 bp on chromosome 6 of goats was most closely related to cashmere traits. The polymorphism of the InDel molecular marker is - / insertion sequence, and the nucleotide sequence of the insertion sequence is shown in SEQ ID NO.1.
Embodiment 2
[0043] Example 2 Identification of the InDel polymorphic site of the Chinese goat FGF5 locus
[0044] 1. Extract the genomic DNA of Pakistani, Nepalese and Chinese goat breeds to be tested
[0045] Among them, there were 146 non-cashmere goats (including 27 non-cashmere goats from Pakistan, 7 Xiangdong black goats from Hunan, 17 Hainan black goats from Hainan, 25 goats from the plains of Nepal, and 7 goats from Shangrao, Jiangxi. 34 Guangfeng goats, 7 Laoshan dairy goats, 10 Tugenbao dairy goats, 19 Yunling black goats); a total of 153 cashmere goats (including 10 Bangor cashmere goats from Tibet, Ritu 10 cashmere goats; 25 Arbas cashmere goats from Inner Mongolia, 4 Alashan cashmere goats, 10 Han cashmere goats; 25 Liaoning cashmere goats from Liaoning; 18 Jining green goats from Shandong, from 30 Southern Xinjiang cashmere goats in southern Xinjiang; 21 cashmere goats from high-altitude areas in Nepal), using conventional methods to extract genomic DNA from blood.
[0046] 2...
Embodiment 3
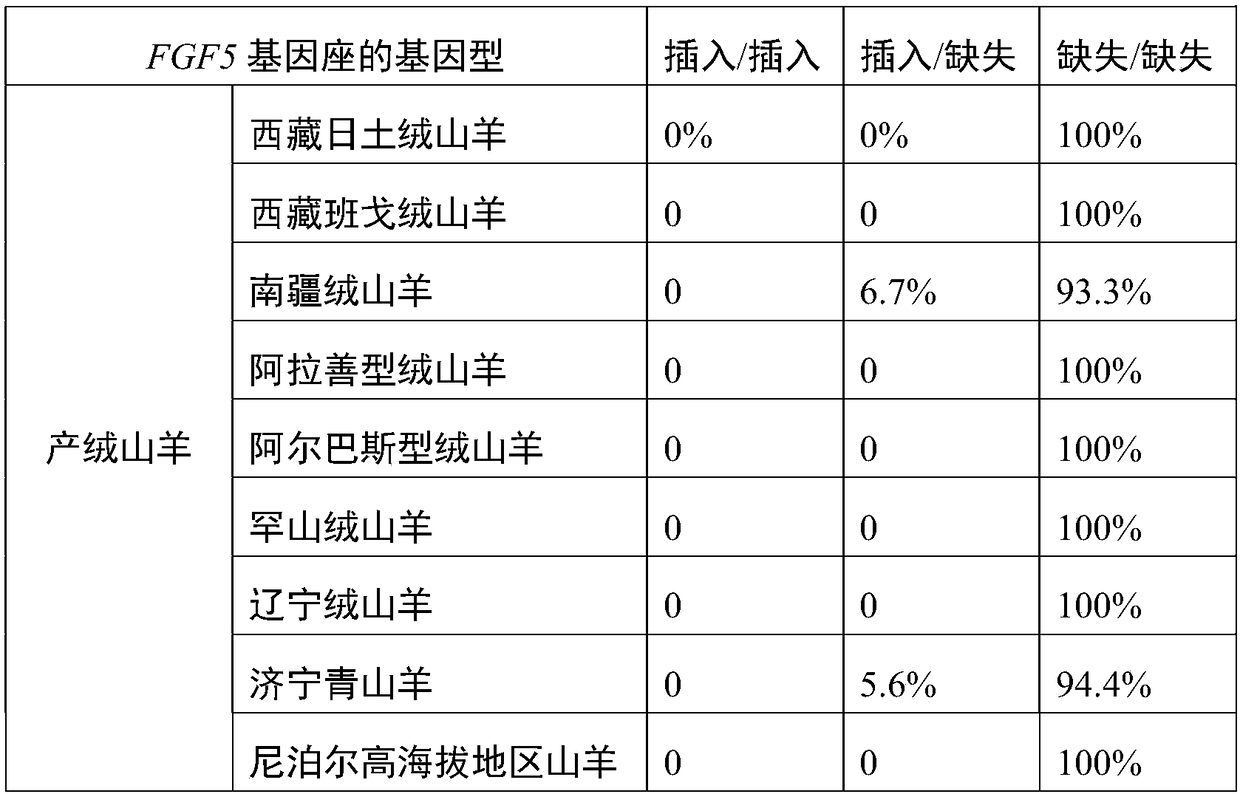
[0051] Example 3 Expanded population analysis of the polymorphic site of the FGF5 gene locus of 299 goats
[0052] According to Example 1, the genomic DNA samples of 153 cashmere-producing goats and 146 non-cashmere-producing goats were extracted, and the specific primers in Example 1 were used for PCR detection, followed by agarose gel electrophoresis, and genotyping based on Sanger sequencing detection. The 153 cashmere goats are Tibetan Bangor cashmere goats, Tibet Ritu cashmere goats, Inner Mongolia Alashan cashmere goats, Inner Mongolia Arbas cashmere goats, Inner Mongolia Han cashmere goats, Liaoning cashmere goats, Nanjiang cashmere goats, Jining green goats 1, Nepal high-altitude goats; 146 non-cashmere goats, namely Pakistan goats, goats in the plains of Nepal, Yunling black goats, Hainan black goats, Guangfeng goats, Hunan black goats, Laoshan milk goats, Tugenbao milk goats goat. Table 1 shows the genotyping results of the FGF5 locus of the FGF5 gene sequence show...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com