Method for rapidly capturing and building database
A fast and linker sequence technology, applied in the field of high-throughput sequencing, can solve the problems of time-consuming, cumbersome steps, and restricting the throughput of library construction, so as to avoid large-scale amplification, increase effective concentration, and save consumption.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
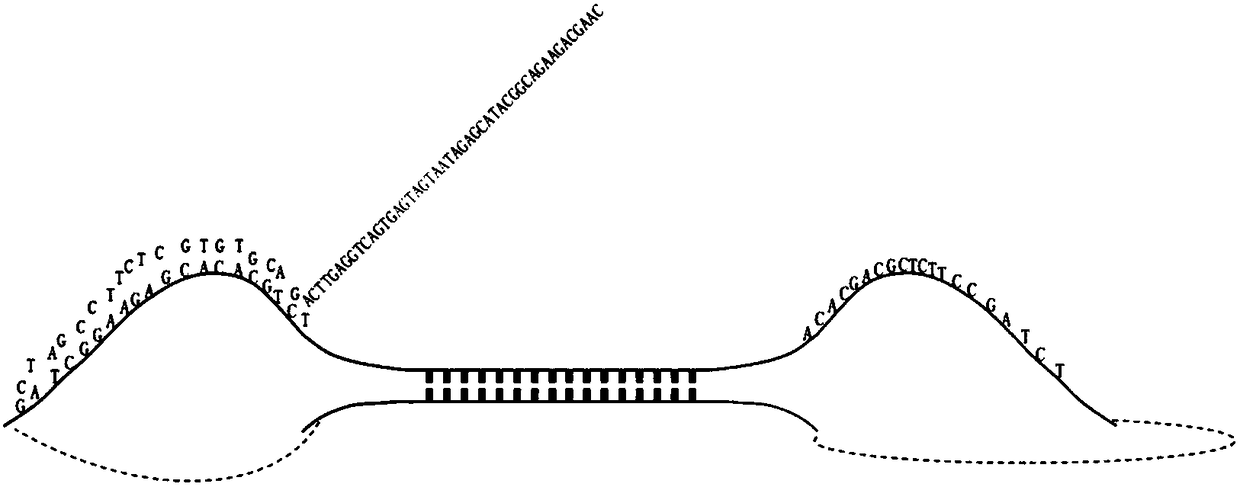
[0075] The extracted genomic DNA was subjected to 3 rounds of fragmentation. After each round, the sample was placed on a shaker to mix well, and after a short centrifugation, the next round of fragmentation was performed.
[0076] Take 1ul sample for capillary electrophoresis detection. After normal interruption, the main peak of the sample detection is about 150bp-200bp.
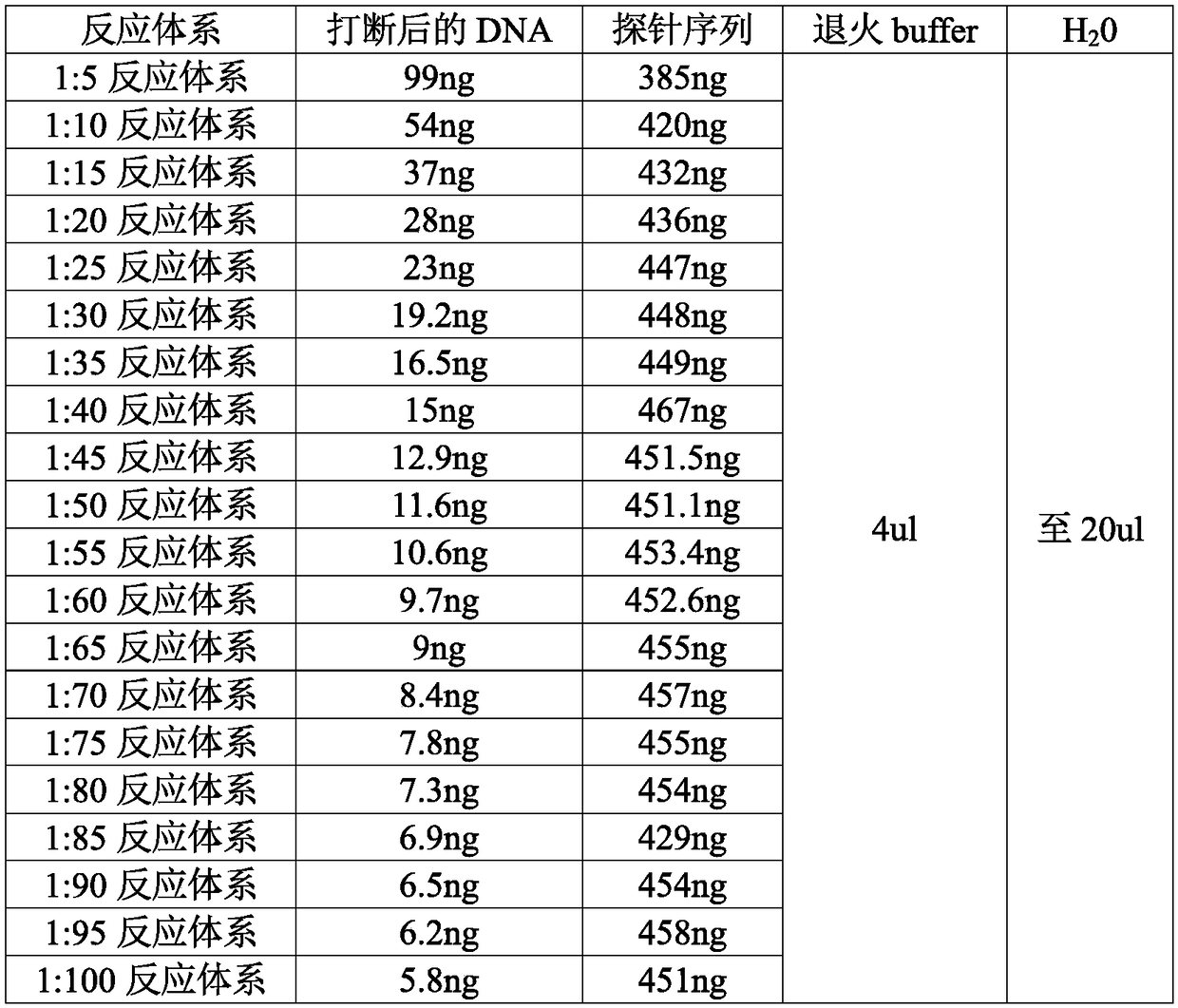
[0077] Annealing: The reaction condition is denaturation at 95°C for 5 minutes, and then annealing. The reaction condition is to drop 0.1°C every 8s to 25°C for 90 minutes. In order to form more rolling circle complexes in the ligation reaction, the ratio of the number of DNA molecules after interruption to the number of probe molecules is designed as follows: 1:5, 1:10, 1:15, 1 :20, 1:25, 1:30, 1:35, 1:40, 1:45, 1:50, 1:55, 1:60, 1:65, 1:70, 1:75, 1:80 , 1:85, 1:90, 1:95, 1:100, and at the same time ensure that the total amount of DNA and probe after fragmentation is 10 pmol.
[0078]
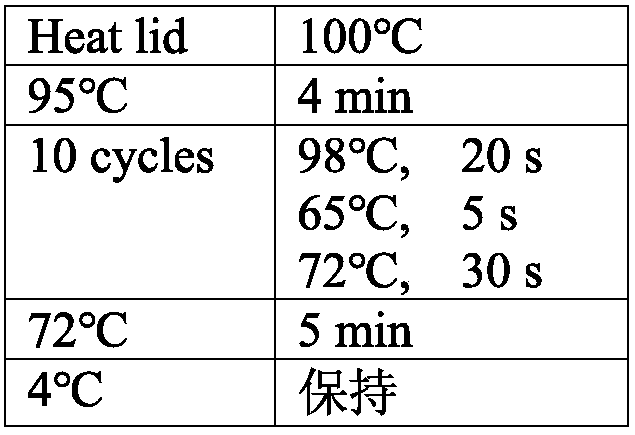
[0079] 1. Con...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com