Ma pomelo InDel (insertion-deletion) molecular marker, and application of molecular marker in differentiating rough bark Ma pomelo at early stage of citrus seed seedling
A molecular marker, Majia pomelo technology, applied in the field of genetics and breeding, can solve problems such as easy mixing, impure seedlings, loss, etc., and achieve the effects of high accuracy, reducing breeding workload and eliminating interference.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] (1) Genomic DNA of fine-skinned Majia pomelo (you series), rough-skinned Majia pomelo, Guanxi honey pomelo, Shatian pomelo and local soil pomelo in Guangfeng County, Jiangxi Province was extracted and prepared by the improved CTAB method.
[0058] (2) Using the primer pair chr2.10-9584F / chr2.10-9584R to amplify and analyze the genomic DNA of all the citrus above. The amplification reaction was carried out in a PTC-200 PCR machine (MJ. Research Inc., USA). The reaction system for PCR amplification is 7μl of Mix solution, 0.3μl of 10mmol / L forward primer, 0.3μl of 10mmol / L reverse primer, 1.5μl of 100ng / ul template DNA and 6μl of ddH 2 O. The specific steps are: denaturation at 95°C for 5 minutes; followed by 35 cycles: 94°C, 30s, 55°C, 30s, 72°C, 35s; finally 72°C, 10min, 12°C, 30min, and 4°C storage.
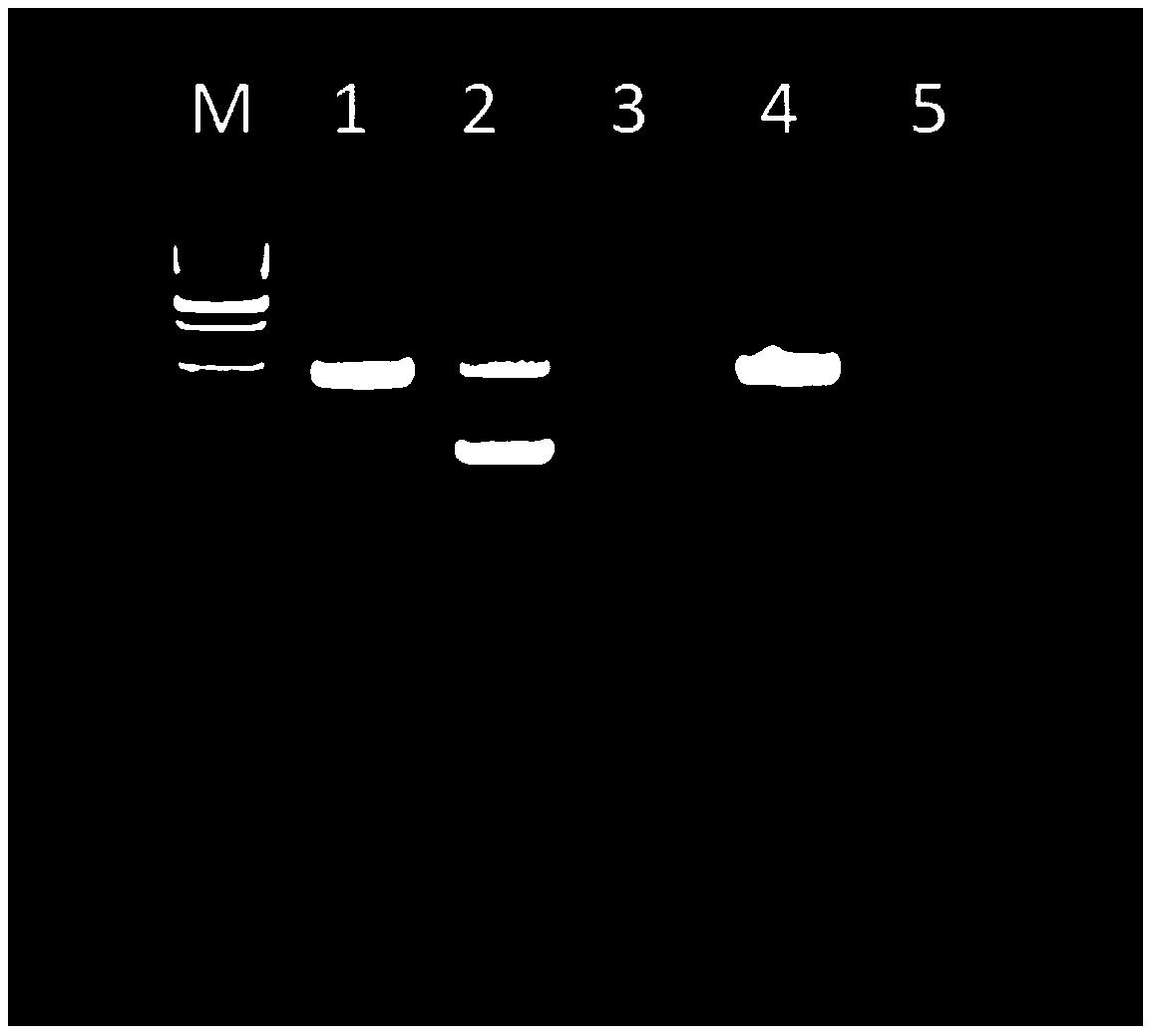
[0059] (3) The amplified product was detected by 3.0% agarose gel electrophoresis on a horizontal electrophoresis tank, using 1×TAE buffer (0.04M Tris-acetate, 0.001M E...
Embodiment 2
[0062] (1) Genomic DNA of Majia pomelo and Guanxi pomelo were extracted and prepared by the improved CTAB method.
[0063] (2) Using the primer pair p-chr6.21-8689F / p-chr6.21-8689R to amplify and analyze the genomic DNA of all the above citrus. The amplification reaction was carried out in a PTC-200 PCR machine (MJ. Research Inc., USA). The reaction system for PCR amplification is 7μl of Mix solution, 0.3μl of 10mmol / L forward primer, 0.3μl of 10mmol / L reverse primer, 1.5μl of 100ng / ul template DNA and 6μl of ddH 2 O. The specific steps are: 95°C denaturation for 5 minutes; followed by 35 cycles: 94°C, 30s, 55°C, 30s, 72°C, 35s; finally 72°C, 10min, 12°C, 30min, 4°C storage.
[0064] (3) The amplified product was detected by 3.0% agarose gel electrophoresis on a horizontal electrophoresis tank, using 1×TAE buffer (0.04M Tris-acetate, 0.001M EDTA, pH8.0), voltage 120V, electrophoresis for 20min. After electrophoresis, the gel imaging system (UVP) took pictures and saved them...
Embodiment 3
[0067] (1) 80 random seedling plants of Majia pomelo (excellent line) mixed with rough skin Majia pomelo were used as test materials, and the genomic DNA of the above test materials was extracted and prepared by the improved CTAB method.
[0068] (2) Using the primer pair p-chr6.21-8689F / p-chr6.21-8689R to amplify and analyze the genomic DNA of all the above citrus. Amplification reactions were performed in a PTC-200 PCR instrument (MJ. Research Inc., USA). The reaction system for PCR amplification is 7μl of Mix solution, 0.3μl of 10mmol / L forward primer, 0.3μl of 10mmol / L reverse primer, 1.5μl of 100ng / ul template DNA and 6μl of ddH 2 O. The specific steps are: 95°C denaturation for 5 minutes; followed by 35 cycles: 94°C, 30s, 55°C, 30s, 72°C, 35s; finally 72°C, 10min, 12°C, 30min, 4°C storage.
[0069] (3) The amplified product was detected by 3.0% agarose gel electrophoresis on a horizontal electrophoresis tank, using 1×TAE buffer (0.04M Tris-acetate, 0.001M EDTA, pH8.0),...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com