Prunus mume weeping trait SNP molecular marker and application thereof
A molecular marker, plum blossom technology, applied in recombinant DNA technology, DNA/RNA fragment, microbial determination/inspection, etc., can solve the problems of low efficiency, long screening cycle of plum blossom weeping traits, etc., achieve good repeatability, improve breeding Efficiency, the effect of shortening the breeding cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1 Based on the SLAF-seq technology, the development of SNP molecular markers for plum blossom weeping traits 1. Experimental materials
[0067] The test materials included 'Liupetal' plum as the female parent, 'Fentai Weeping Branch' as the male parent and F 1 Separate groups, straight and weeping plum varieties. The parents had significant differences in phenotypic traits such as weeping branch and flower color, and all the materials were planted in He Village, Moganshan Town, Huzhou City, Zhejiang Province (30.566389°N, 119.879582°E).
[0068] 2. Genomic DNA extraction
[0069] choose F 1 There are 20 individual plants of straight branch and weeping branch plant type in the group. DNA extraction was carried out according to the instructions of the high-efficiency plant genomic DNA extraction kit (Tiangen Biochemical Technology Co., Ltd.), and 1.0% agarose gel was prepared, and 3 μL DNA was mixed with about 1 μL loading-buffer, and the voltage was 150 V, a...
Embodiment 2
[0072] The amplification of embodiment 2 SNP molecular marker
[0073] Primers were designed according to the differential markers of plum blossom weeping characters, as follows:
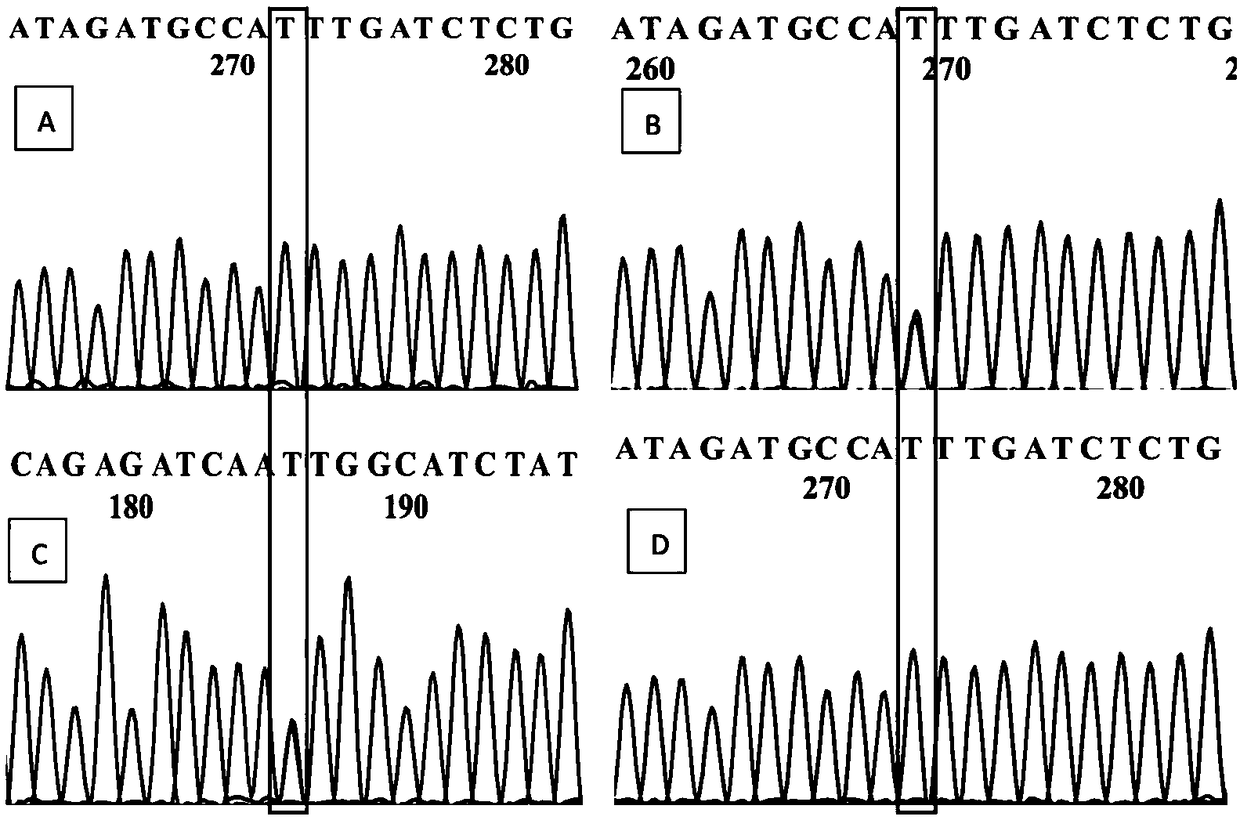
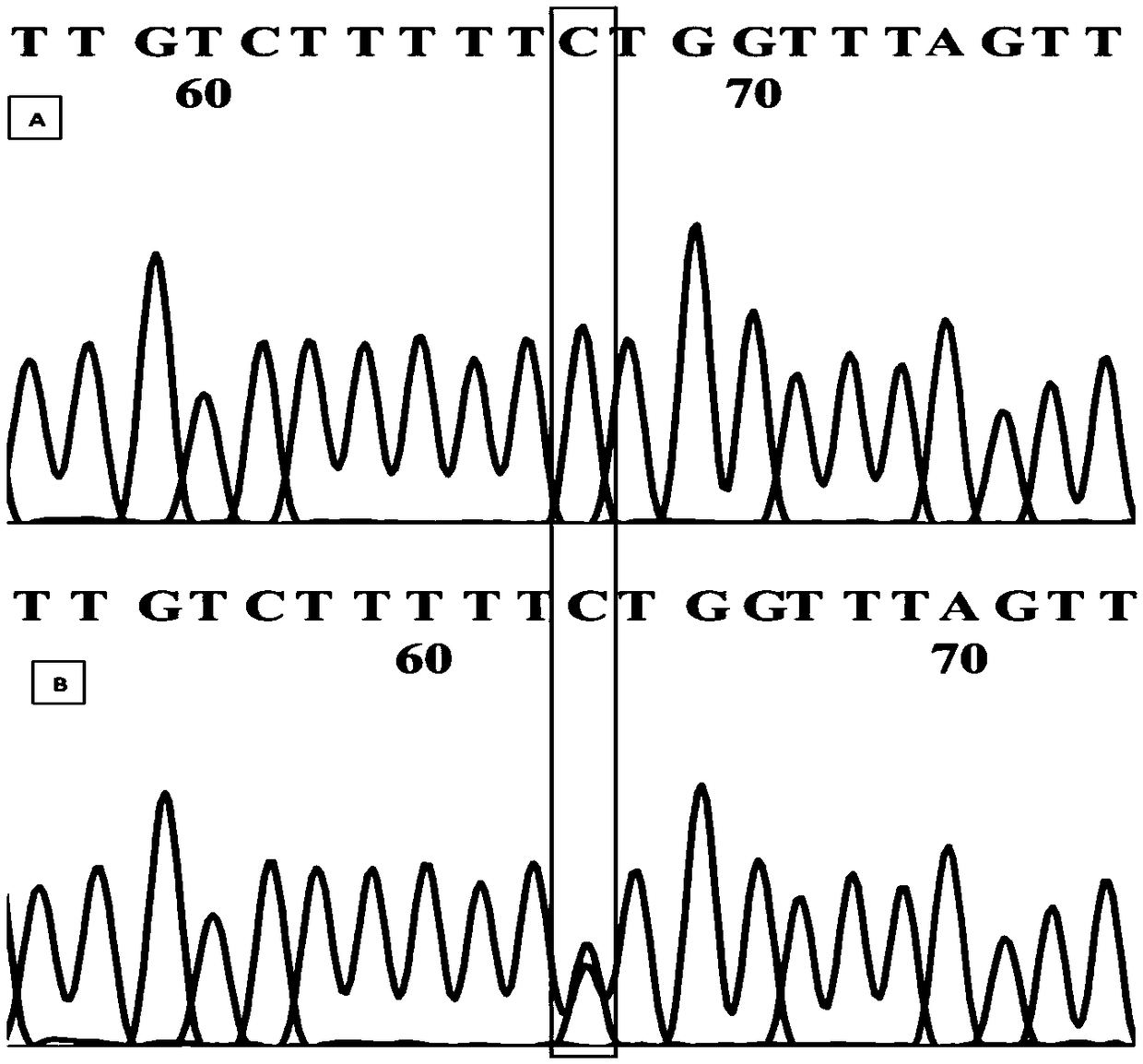
[0074] According to the Sanger sequencing results, two Markers (Marker301243, Marker311414) were selected, and primers were designed using Primer Premier 5.0. The primer sequences are shown in Table 1.
[0075] Table 1 Information of 2 pairs of primers used for Sanger sequencing
[0076]
[0077] Several straight and weeping branches were randomly selected for PCR amplification. PCR amplification system 20μL, containing 100ng·μL -1 Template DNA 2 μL, 2×Taq PCR Master Mix (BIOMIGA) 10 μL, 10 mol / L forward and reverse primers (Beijing Synthesis Department of Sangon Bioengineering Co., Ltd.) 1 μL each, ddH 2 O 10.3 μL. The PCR program was: pre-denaturation at 95°C for 2 min; denaturation at 94°C for 20 s, annealing at 56°C for 30 s, extension at 72°C for 20 s, and 30 cycles; final extension at 7...
Embodiment 3
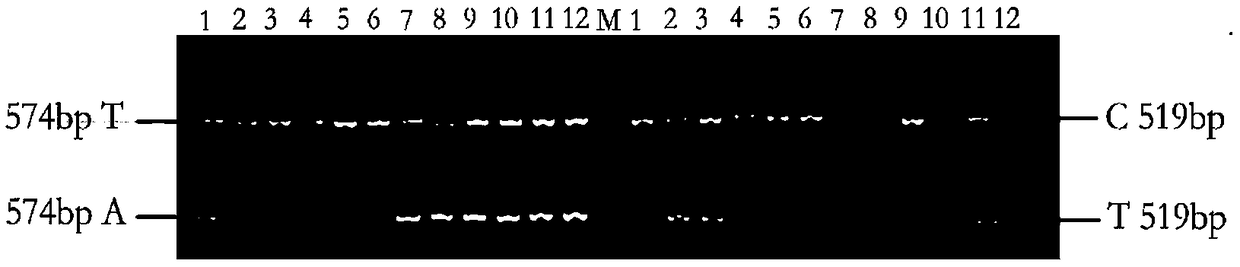
[0079] Example 3 Establishing an AS-PCR method for genotyping based on SNP markers
[0080] Two markers (Marker301243 and Marker311414, whose nucleotide sequences are shown in SEQ ID NO.1 and SEQ ID NO.2 respectively) were obtained by Sanger sequencing, and AS-PCR primers were designed for corresponding SNP sites. When designing allele-specificity, the SNP sites of Marker301243(A / T) and Marker311414(C / T) were designed respectively. In order to improve the specificity of primers, at the same time, introduce 1 mismatched base, another primer (common primer) was designed according to conventional methods, and the specificity of the set primer was detected by NCBI (Table 2). During the AS-PCR operation, two AS-PCR primers should be paired with common primers, and the same DNA template should be amplified by PCR in two PCR tubes. If the 3' end of the AS-PCR primer matches the allelic site in the sample, effective amplification can be performed; otherwise, effective amplification can...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com