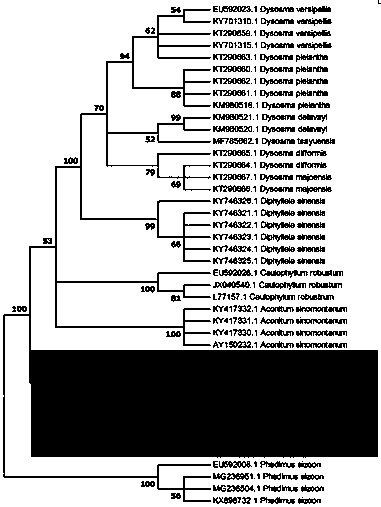
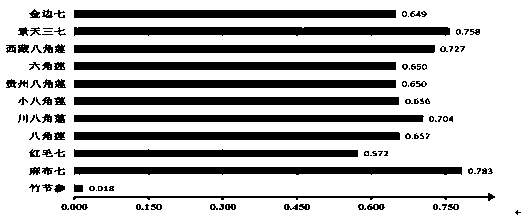
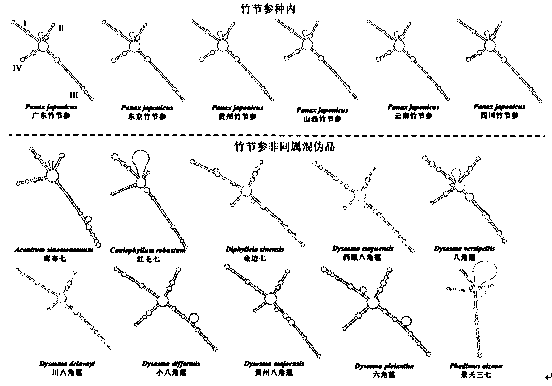
Method for identifying Japanese ginseng rhizomes and non-generic confusion or counterfeit species of Japanese ginseng rhizomes based on ITS2
A technology of bamboo ginseng and counterfeit products, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as the complexity of bamboo ginseng, achieve simple and easy identification, accurate results, and avoid singleness Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] 1. DNA extraction (Plant Tissue DNA Rapid Extraction Kit Fatiangen DP305), as follows.
[0022] (1) The dry weight tissue is about 30 mg, which is fully ground by adding liquid nitrogen.
[0023] (2) Quickly transfer the ground powder to a centrifuge tube pre-filled with 700 µl 65°C preheated buffer GP1 (add mercaptoethanol to the preheated GP1 before the experiment to make the final concentration 0.1%), and quickly After mixing by inversion, the centrifuge tube was placed in a 65°C water bath for 20 min, and the centrifuge tube was inverted several times during the water bath to mix the samples.
[0024] (3) Add 700 µl of chloroform, mix well, and centrifuge at 12,000 rpm (~13,400×g) for 5 min. Note: If the plant tissue rich in polyphenols or starch is extracted, an equal volume extraction can be performed with phenol:chloroform / 1:1 before step 3.
[0025] (4) Carefully transfer the upper aqueous phase obtained in the previous step into a new centrifuge tube, add 70...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com