Industrial strain and method for efficient xylose metabolism based ethanol production
A technology for xylose metabolism and ethanol production, applied in the direction of microorganism-based methods, biochemical equipment and methods, fungi, etc., can solve the problems of Saccharomyces cerevisiae toxicity and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 Effects of single copy, secondary copy, and triple copy of the ethanol-producing gene expression cluster of haploid Saccharomyces cerevisiae on ethanol production by yeast fermentation
[0027] 1. Gene expression clusters of ethanol production in Saccharomyces cerevisiae
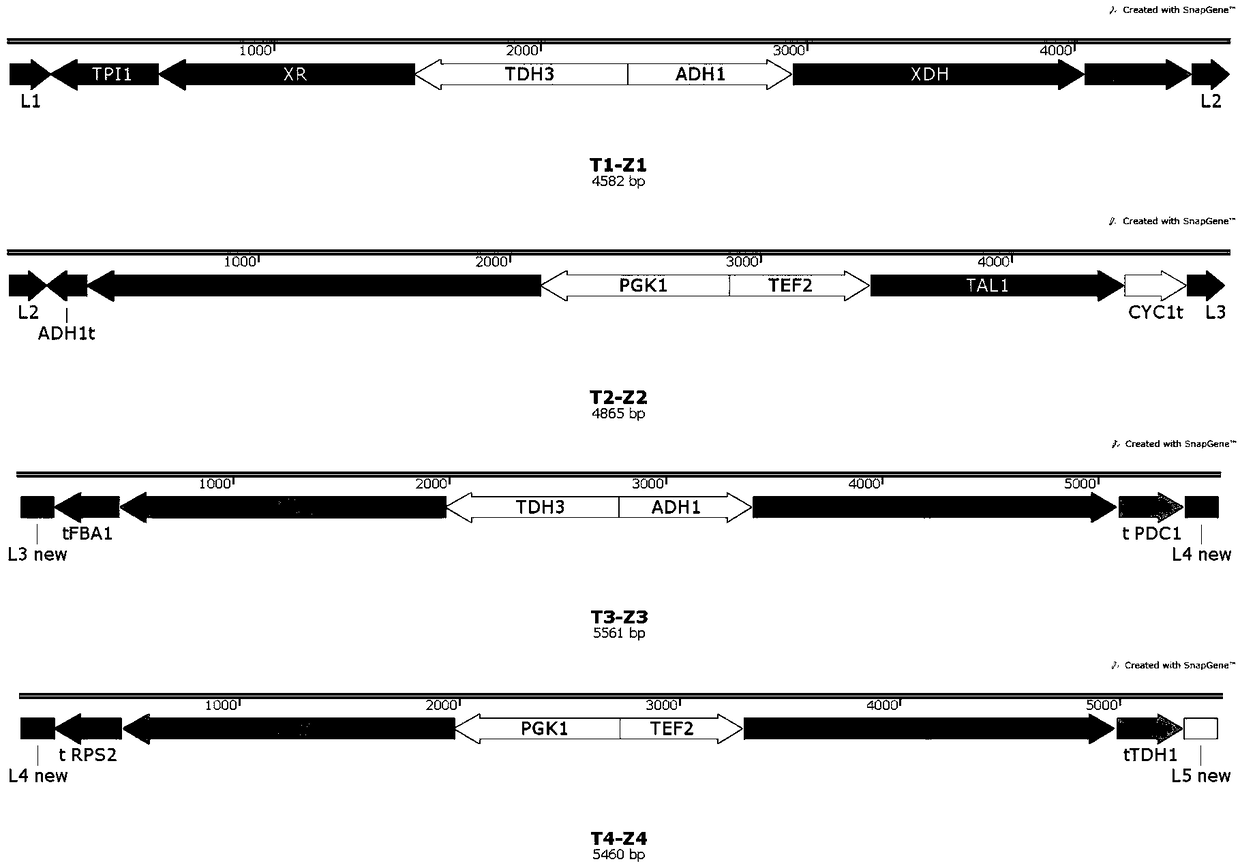
[0028] Select xylose reductase XR, xylitol dehydrogenase XDH, xylulokinase XK, and xylose transporter genes mgt05196, TAL1, PYK1, and select constitutive strong promoters pPGK1, pADH1, pTDH3, pTEF1 to express the above Six genes, PYK1, TAL1, XR, XDH, XK, and mgt05196, were integrated into the S. cerevisiae genome.
[0029] In order to realize the co-transfer of 6-8 genes into the yeast genome, the selected vector contains the integration site (L1-L6) and the expression fragment (the endogenous terminator of S. For homologous recombination between expression modules, since two Saccharomyces cerevisiae terminators that are reversed to each other are already contained, the cause of plasmid con...
Embodiment 2
[0052] Example 2 Effect of random multiple copies of haploid Saccharomyces cerevisiae gene expression cluster on ethanol production by yeast fermentation
[0053] The expression clusters in Example 1: T1-Z1, T2-Z2, T3-Z3, T4-Z4 (including XR (K270R), XDH, XK, TAL1, PYK1 (2 copies), mgt05196 (2 copies) ) was integrated into 1Z1Z bacteria in the form of DELTADNA or rDNA-mediated random multiple copies, so as to explore the effect of multiple copies of the expression cluster on the fermentation results of Saccharomyces cerevisiae.
[0054] DELTA7 and DELTA9 are two random multi-copy strains mediated by DELTA DNA, rDNA8 and rDNA12 are two random multi-copy strains mediated by rDNA.
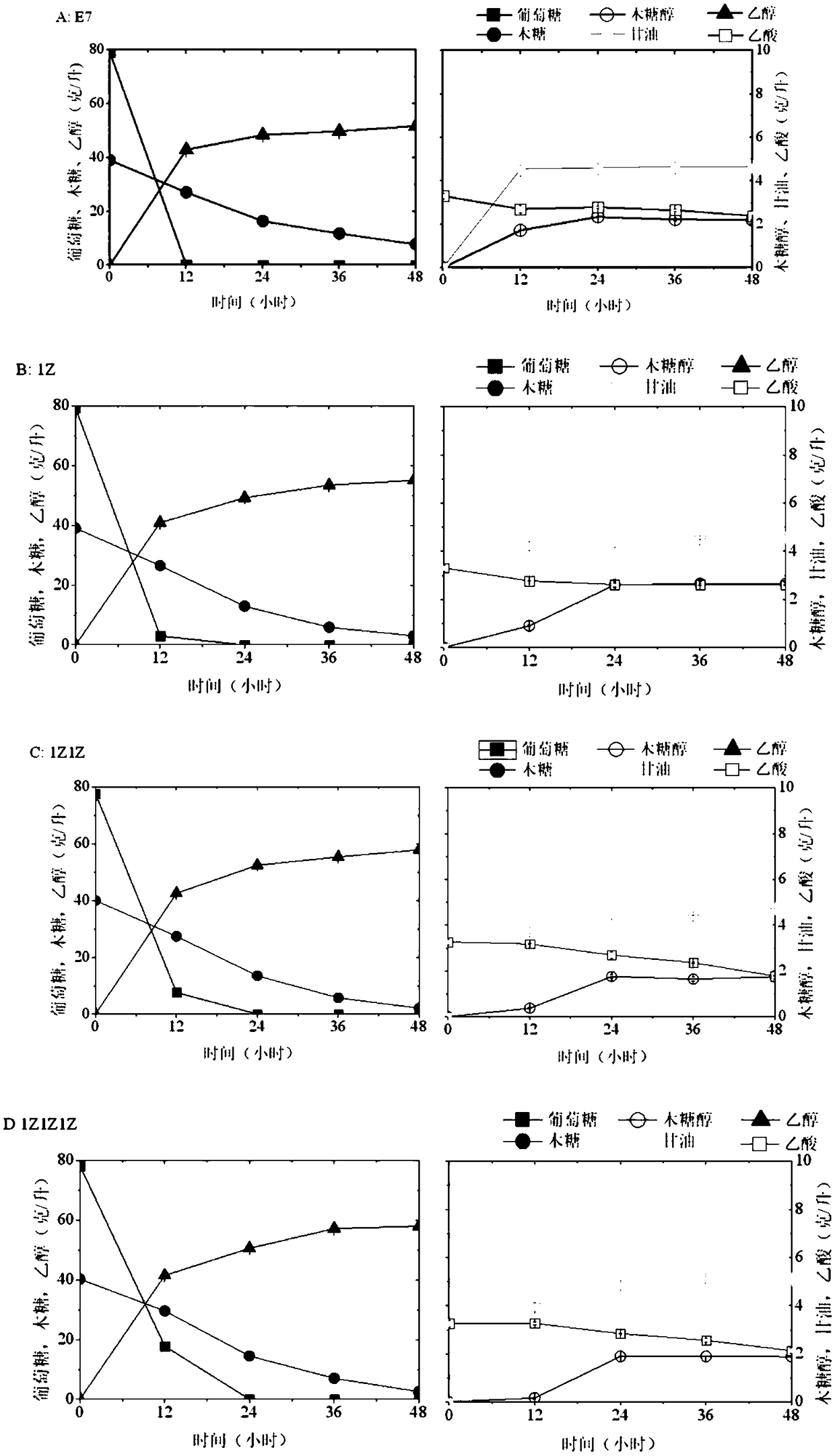
[0055] The fermentation results of the initial strain 1Z1Z and DELTA7, DELTA9, rDNA8, rDNA12 are as follows Figure 4 :
[0056] After screening the resistant strains on the AbA resistance plate and verifying the correct sequence by PCR cloning, a total of 8 strains were screened out as positive clo...
Embodiment 3
[0061] Example 3 Transcription level analysis of haploid yeast random multi-copy gene expression clusters
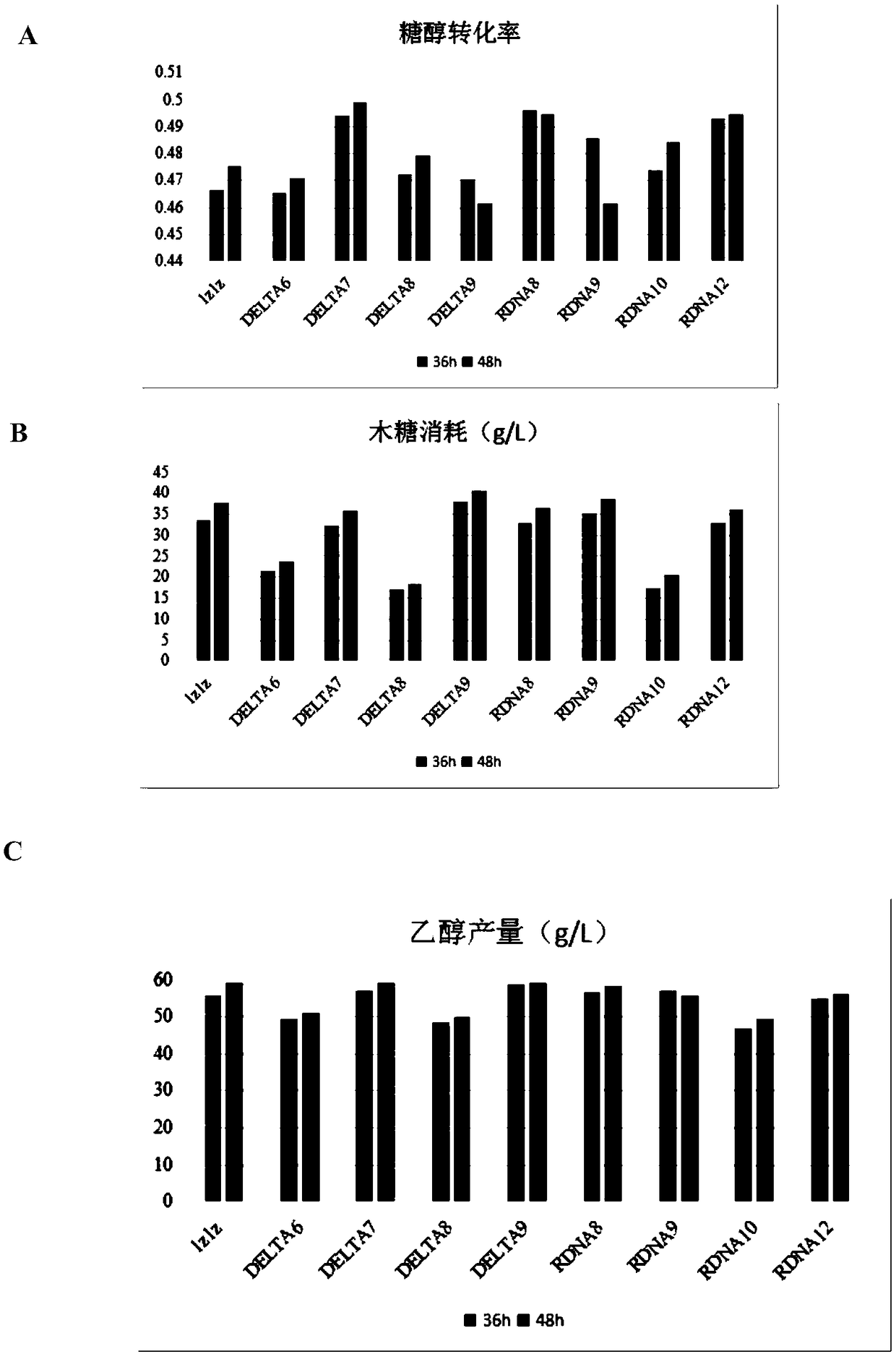
[0062] When the gene expression cluster is integrated and expressed in the yeast genome in the form of random multiple copies, not only does not appear that the fermentation performance of the gene expression cluster in the third copy is not much different from that of the second copy, but it shows a high ethanol fermentation performance. The differences in the transcription levels of six genes XR(K270R), XDH, XK, TAL1, PYK1 and mgt05196 among the four strains of DELTA7, DELTA9, rDNA8 and rDNA12 were detected, so as to determine the reason for the improvement of the ethanol fermentation performance of the bacteria. The following are the multiples of transcription levels of the six genes XR (K270R), XDH, XK, TAL1, PYK1, and mgt05196 of the four strains DELTA7, DELTA9, rDNA8, and rDNA12 compared to the original strain 1Z1Z, such as Figure 5 .
[0063] The results showed...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com