Method for identifying genotype of insertion deletion locus of animal gene with HRM
An insertion-deletion and site-based technology, applied in the fields of cell engineering and molecular biology, can solve the problems of long identification time, difficult experimental operation, high cost, etc., and achieve the effect of low application cost, high accuracy and fast speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
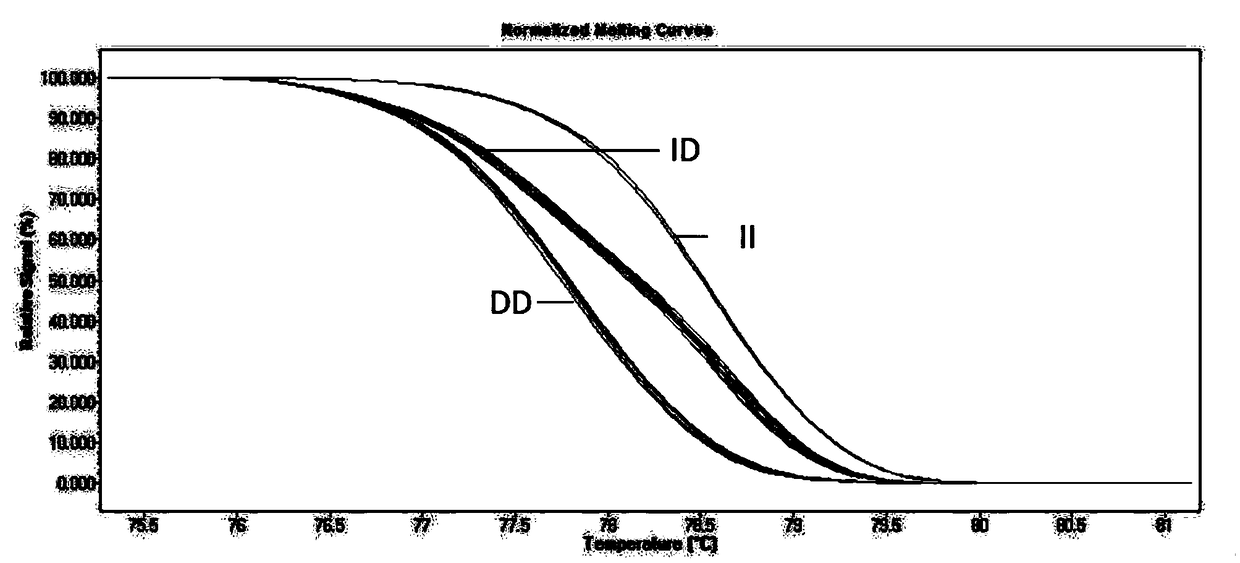
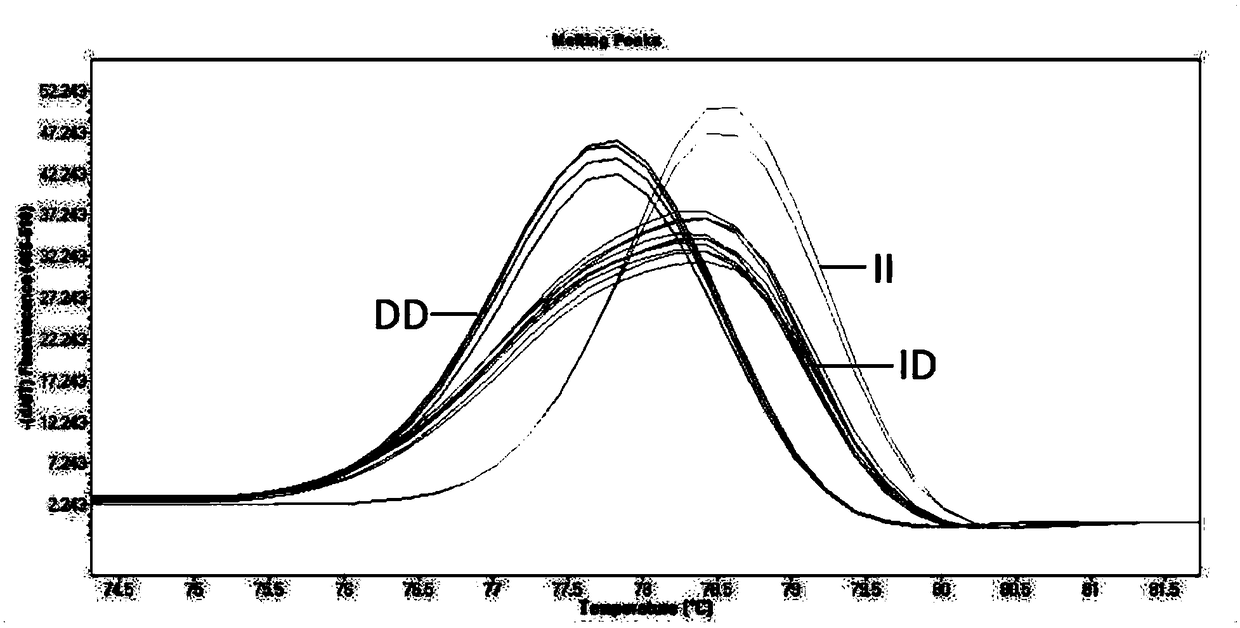
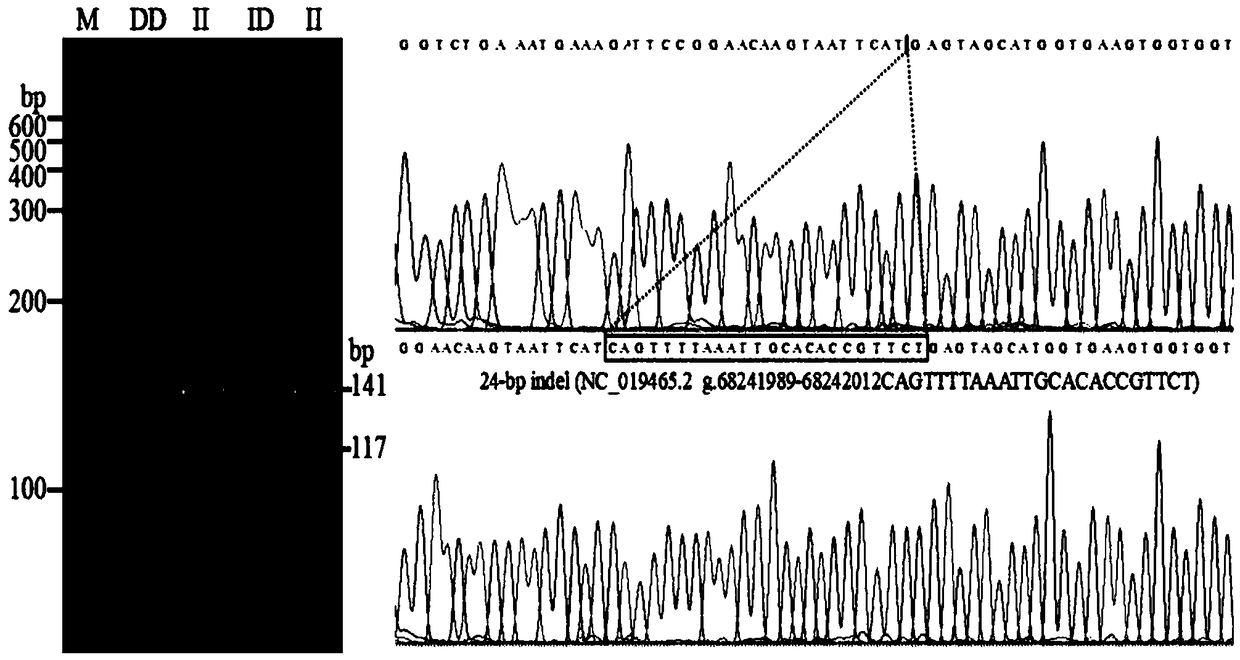
[0066]Example 1: Identification of indel sites by HRM of PLAGL1 gene in 7 sheep breeds
[0067] Collection of sheep blood samples and ear tissue samples: 262 samples of small-tailed Han sheep were collected from Lanzhou City, Gansu Province; 166 samples of the same sheep were collected from Baishui County, Shaanxi Province; 67 samples of Lanzhou Big-tailed sheep were collected from Lanzhou City, Gansu Province , 200 samples of Hu sheep, collected from Huzhou City, Zhejiang Province, 96 samples of Duolang sheep, collected from Shihezi City, Xinjiang, 93 samples of Bashibai sheep, collected from Shihezi City, Xinjiang, 92 samples of Altay sheep, collected from Xinjiang In Altay City, a total of 976 sheep samples were collected, and all samples were transported on dry ice and stored in a -80°C refrigerator.
[0068] Sheep sample DNA extraction work:
[0069] Using the Whole Blood DNA Extraction Kit of Adelaide Biotechnology Co., Ltd. Operation steps: Take 200 microliters of fres...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com