Recombinant plasmid for production of oxalate decarboxylase, system and method for expressing escherichia coli, and application
A technology of oxalate decarboxylase and Escherichia coli, applied in the biological field, can solve the problems of complex renaturation process, high cost, inactivity and the like, and achieve the effects of simple separation and purification process, low production cost and high specific activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
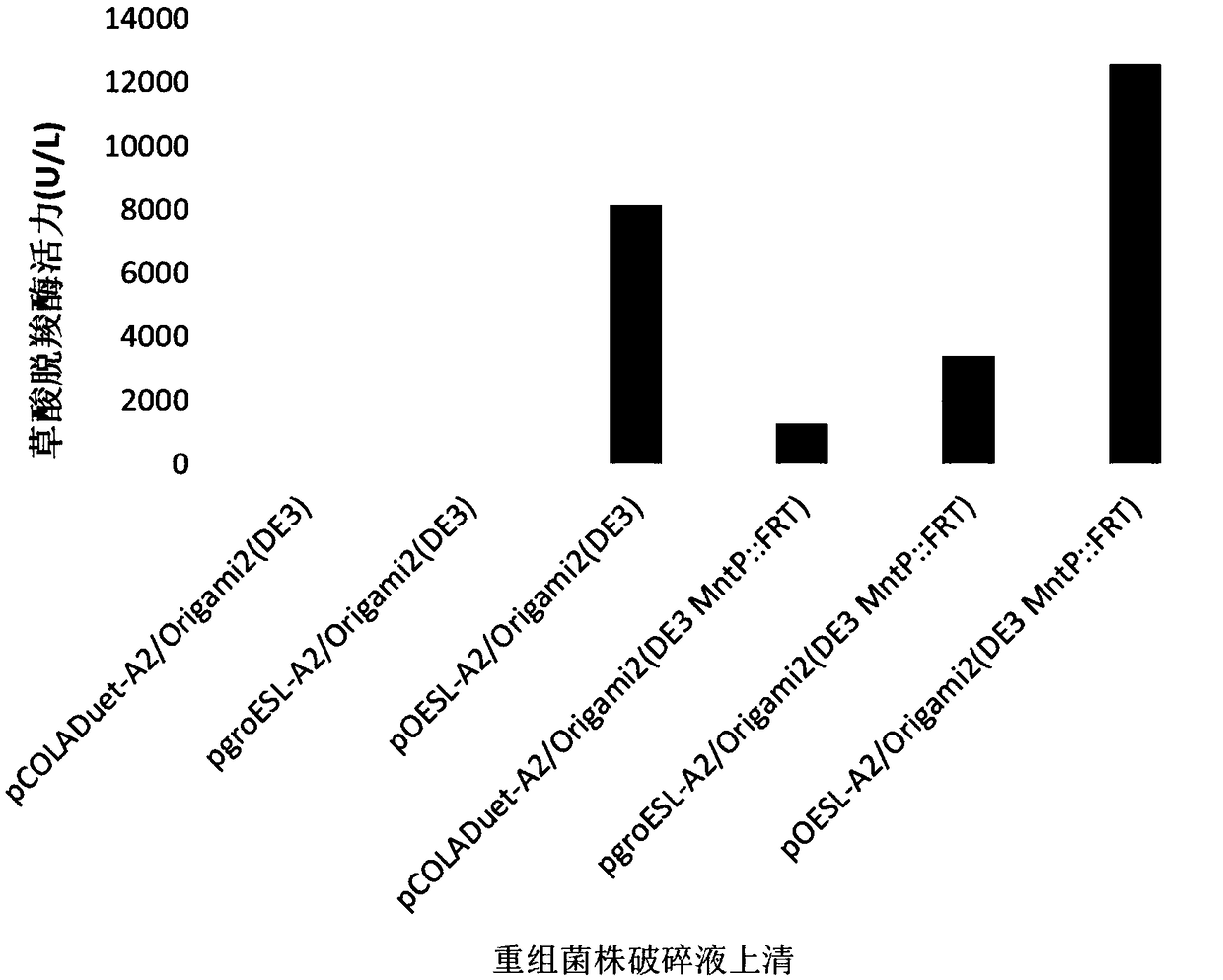
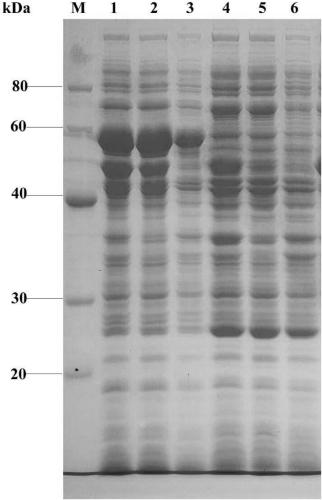
[0043] The construction of embodiment 1 expression vector pCOLADuet-A2
[0044] Using the oxalate decarboxylase A2 gene (as shown in the sequence table SEQ ID NO.1) synthesized by the whole gene as a template, design the primer pair F1 / R1, amplify the gene, and perform gel recovery and purification on the amplified product. The method refers to According to the method described in the instructions of the commercially available DNA mini-purification kit, the A2 gene fragment was finally obtained. PCR system: 10×PCR Buffer 5μL, 2mMdNTP 5μL, 25mM MgSO 4 5 μL, 1.5 μL each of 10 μM primer F / R, 0.5 μL template DNA, 1 μL KOD-Plus-Neo, ddH 2 O32.5μL; PCR reaction conditions are as follows: 94°C for 3min, 30 cycles (98°C for 10s, 60°C for 30s, 68°C for 35s), 68°C for 5min, 4°C for 10min; They are all the same, and will not be described in detail below. The PCR reaction conditions are slightly different, mainly due to the difference in annealing temperature and extension time. Extra...
Embodiment 2
[0047] The construction of embodiment 2 expression vector pgroESL-A2
[0048]Using the recombinant plasmid pCOLADuet-A2 obtained in Example 1 as a template, design primer F2 / R2 to amplify, and obtain DNA fragment 1 after product purification; (shown) fragment as a template, design primers to amplify F3 / R3, obtain DNA fragment 2 after product purification; use molecular chaperone plasmid pGro7 plasmid as template, design primers to amplify F4 / R4, obtain DNA fragment 3 after purification ; Prepare the following reaction system in an ice-water bath according to the instructions of the seamless cloning kit, connect the above DNA fragments 1, 2 and 3, and transform Escherichia coli DH5α.
[0049] wxya 2 o
Up to 20μl
5×Buffer
4μl
DNA Fragment 1
80ng
DNA Fragment 2
40ng
DNA fragment 3
60ng
2μl
[0050] Use the Inoue method to prepare DH5α supercompetent, the method refers to "Molecular Cloning Exp...
Embodiment 3
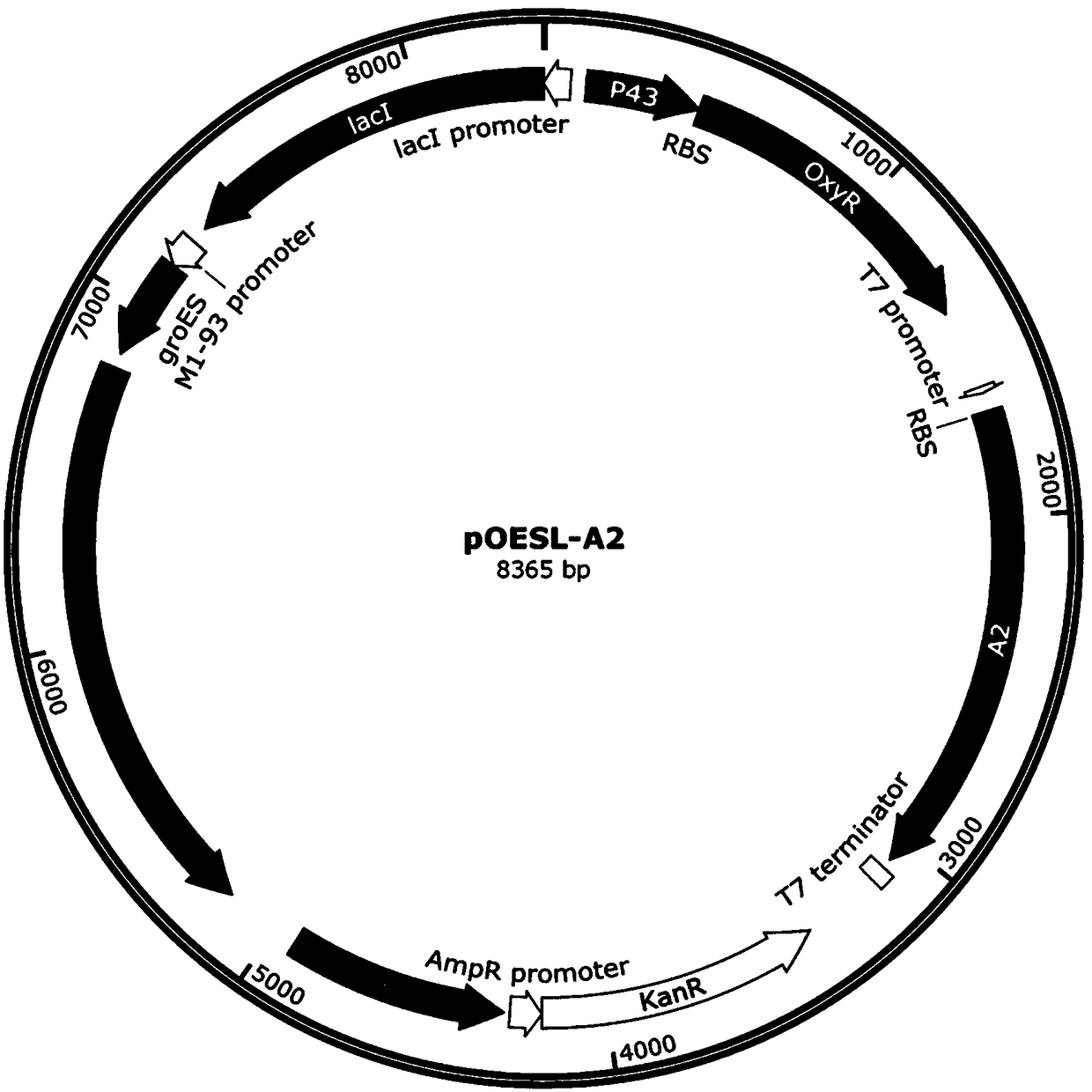
[0057] Example 3 Construction of expression vector pOESL-A2
[0058] Using the genomic DNA of Escherichia coli K12MG1655 strain as template, design primer pair F5 / R5 to amplify OxyR gene and its terminator DNA fragment (as shown in sequence table SEQ ID NO.3); Extract recombinant plasmid pgroESL-A2, use restriction Endonucleases NcoI and EcoRI respectively double-digested the DNA fragments of the OxyR gene and terminator and the pgroESL-A2 plasmid, ligated the above-mentioned double-digested fragments according to the method of the DNA ligase instruction manual, and added 2 μl of the ligated recombinant plasmid to Add 100 μl of DH5α competent cells, place in an ice-water bath for 30 minutes, heat shock in a 42°C water bath for 90 seconds, place in ice for 3 minutes, add 800 μl of antibiotic-free medium, and incubate on a constant temperature shaking shaker at 37°C for 40 minutes. Take 100 μl of the resistant LB solid medium plate containing 50 μg / ml kanamycin for screening, an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com