Identification and Application of an Enhanced Gene Expression Sequence in Brassica napus
A technology in sequences and sequence listings, applied in the field of identification of enhanced gene expression sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
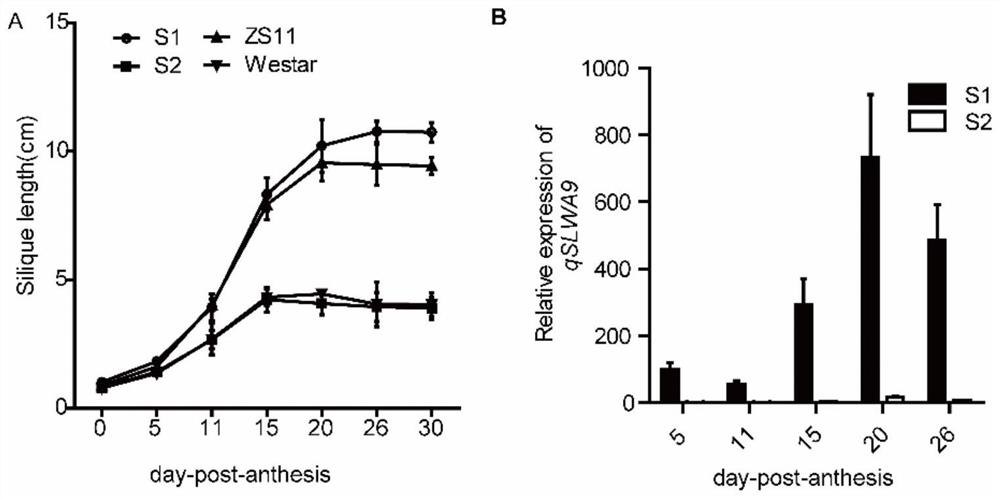
[0030] Example 1: Drawing of silique growth curve and expression level of qSLWA9 gene at different developmental stages
[0031] In order to visually compare the growth of siliques in the whole development period of Brassica napus, the applicant selected a Brassica napus variety Westar (a known and public variety) with common silique length (about 5-6cm) and thousand-grain weight and production The widely used silique and large-grain variety Zhongshuang 11 (ZS11) was used as a reference, and the growth dynamics of the siliques of S1, S2, ZS11, and Westar were investigated in the experimental field of Huazhong Agricultural University during the spring flowering stage of 2014, and the main inflorescence of the plants was marked. The flowers that bloom on the same day are recorded as 0dpa, and the lengths of at least 8 stigmas or siliques are measured for each material in each period, and the average length is taken as the length of the siliques in this period and the growth curve...
Embodiment 2
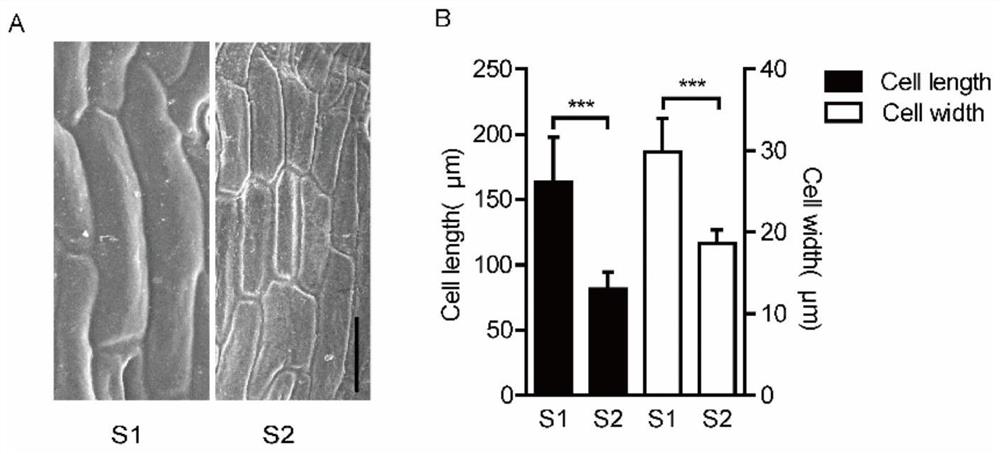
[0033] Example 2: Scanning electron microscope analysis of silique peel cells
[0034] When the siliques were no longer elongated, the siliques were cut into small pieces from the field, fixed with 2.5% glutaraldehyde (prepared with 0.1M PBS), and taken back to the laboratory, vacuumed for 30 minutes, and kept overnight at 4°C. Low alcohol dehydration at different levels, the alcohol concentration is 30%, 50%, 70% for 10 minutes each, 90%, 100% for 8 minutes each, after the dehydration is completed, pour off the alcohol, replace it with isoamyl acetate for 20 minutes, and then carry out the sample Critical point drying (CPD 020, Balzers Union), followed by sample gold spraying (Nanotech SEMPrepⅡsputtercoater), and then taking pictures with a JSM-6390 scanning electron microscope, and the operation of the fixed sample was carried out on the public electron microscope platform of Huazhong Agricultural University. By scanning electron microscope observation of S1 and S2 silique p...
Embodiment 3
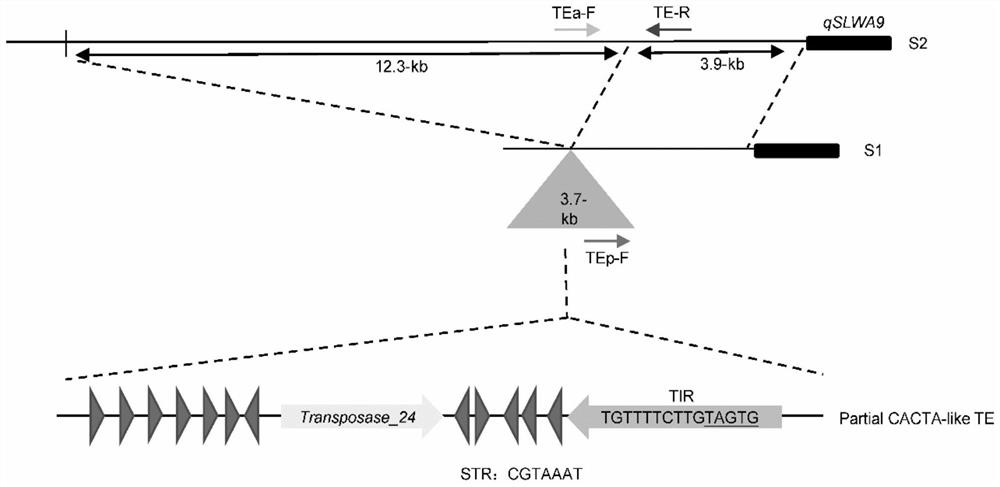
[0035] Example 3: Sequence analysis of the 3.7-kb upstream regulatory region of the silique parent S1 gene qSLWA9 inserted
[0036] Comparative sequencing of the parents showed that a 3.7-kb DNA fragment was inserted 3.9-kb upstream of the qSLWA9 start codon (ATG) of S1, and the sequence was put into the NCBI database (https: / / www.ncbi.nlm.nih .gov / ) and RepeatMasker website (http: / / www.repeatmasker.org / ), and found that the sequence is an incomplete CACTA-like transposon sequence, which contains the right terminal repeat sequence (TIR) (TGTTCCTTG TAG TG ), a transposase family (Transposase_24) gene and subterminal repeat (STR) (CGTAAAT) (see image 3 ).
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com