A kind of pCasPA/pACRISPR two-plasmid system and its application
A plasmid, recombinant system technology, applied in DNA preparation, recombinant DNA technology, introduction of foreign genetic material using vectors, etc., to achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1: pCasPA plasmid construction:
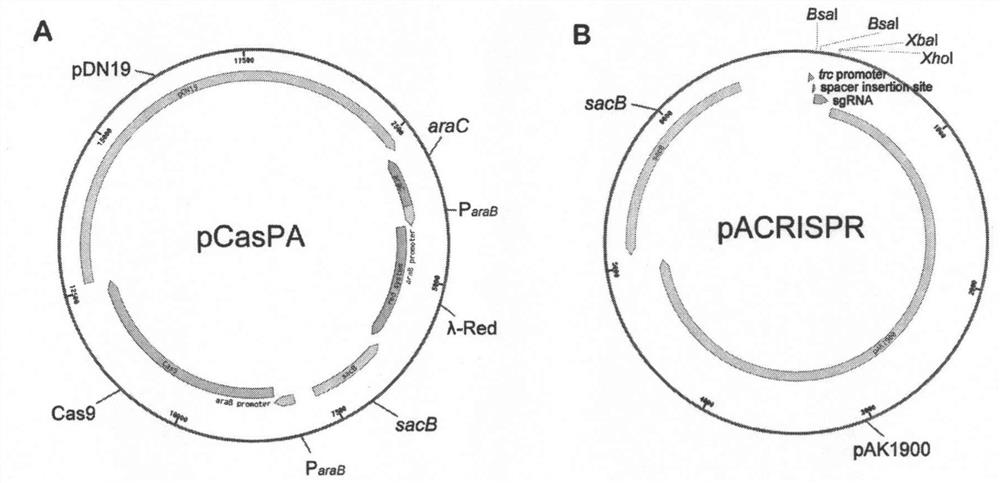
[0042] The composition of the pCasPA plasmid is attached figure 1 As shown in A, its sequence is SEQ ID NO:1. The specific construction method of pCasPA plasmid is as follows:
[0043] (1) Using the pKD46 plasmid as a template, the DNA fragment of the λ-Red recombination system and the arabinose-induced P araB The DNA fragment of the promoter; the gene fragment containing the Cas9 protein was amplified by PCR from the pCasSA plasmid.
[0044] The primer sequences used for PCR amplification are:
[0045] Amplified λ-Red recombination system 5' primer sequence (SEQ ID NO: 3):
[0046] 5'-AACGACGGCCAGTGAATTCGAGCTCGGTACCctttcctgcgttGTCGAC
[0047] tgtatttgaaaaataaacaaataggggtcc-3'
[0048] Amplified λ-Red recombination system 3' primer sequence (SEQ ID NO: 4):
[0049] 5'-gtatgaaaagtCTCGAGCATTCTAGAtccacaGCGGCCGCcatggattcttcgtctgtttctactg-3'
[0050] Amplified P araB Promoter 5' primer sequence (SEQ ID NO: 5):
[0051] 5...
Embodiment 2
[0072] Embodiment two: pACRISPR plasmid construction:
[0073] The composition of the pACRISPR plasmid is attached figure 1 As shown in B, its sequence is SEQ ID NO:2. The specific construction method of pACRISPR plasmid is as follows:
[0074] (1) Digest the pAK1900 plasmid with BamHI-HF and HindIII-HF. The reaction system is as follows: 5 μl 10×CutSmart Buffer, 2 μg pAK1900 plasmid, 2 μl BamHI-HF (20 units / μl), 2 μl HindIII-HF (20 units / μl ), and finally add an appropriate amount of ddH 2 O to a total volume of 50 μl. The buffer and enzymes used in this reaction were produced by NEB Company. After the reaction system was reacted at 37° C. for 2 to 3 hours, the double-digested product was purified and recovered using the SanPrep Column PCR Product Purification Kit produced by Sangon Bioengineering (Shanghai) Co., Ltd. The specific steps were carried out according to the operation manual of the kit.
[0075] (2) Using the pEX18Ap plasmid as a template, the DNA fragment o...
Embodiment 3
[0088] Example 3: The pCasPA / pACRISPR dual plasmid system realizes efficient gene knockout in Pseudomonas aeruginosa strains:
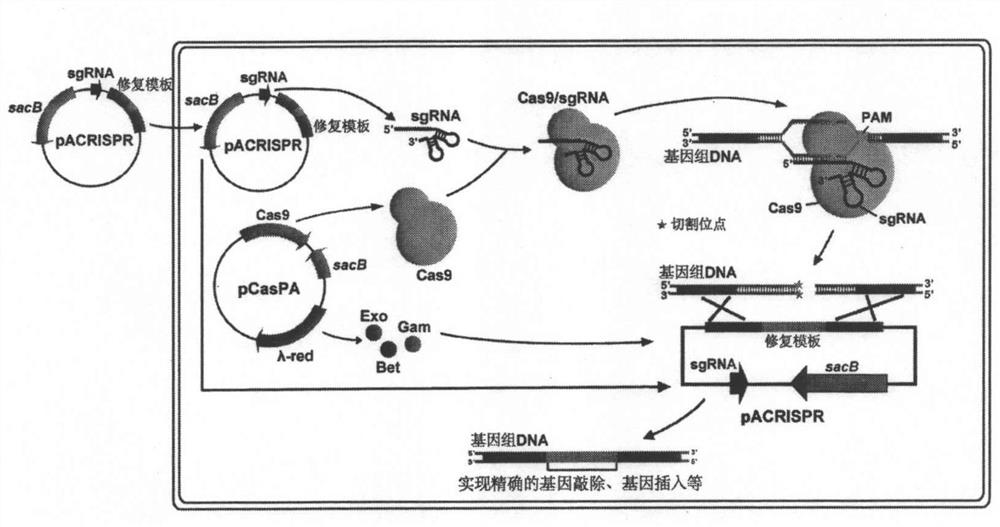
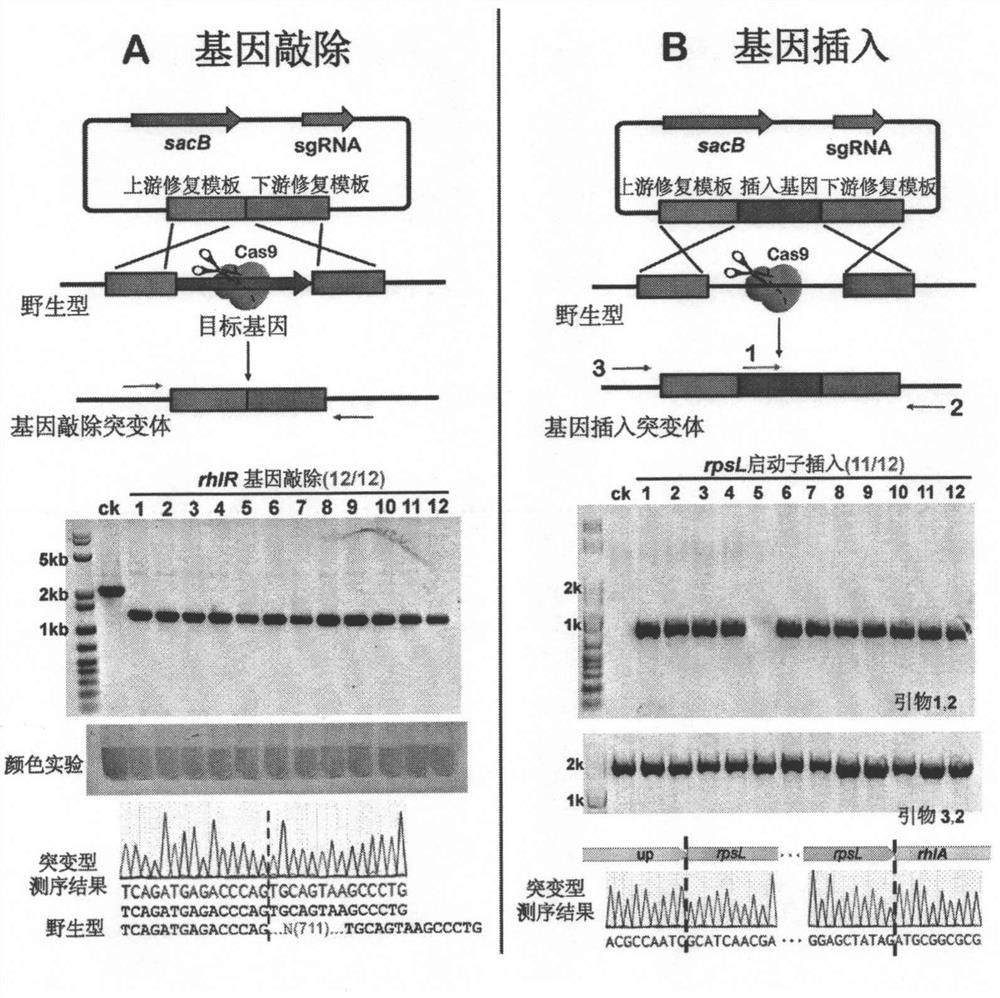
[0089] Using the pCasPA / pACRISPR dual plasmid system can achieve efficient knockout of different genes in various strains of Pseudomonas aeruginosa. The process is shown in the attached figure 2 shown. In the experiment, the rhlR gene was selected as an example, and the gene knockout experiment was carried out in Pseudomonas aeruginosa PAO1 strain. attached image 3 A is a schematic diagram of gene knockout in Pseudomonas aeruginosa using pCasPA / pACRISPR dual-plasmid system, and the results of PCR verification, pigment experiment and DNA sequencing of rhlR gene knockout in PAO1 strain.
[0090] (1) First select a DNA fragment of the first 20 bases of a certain NGG (N is any base) sequence on the target gene of the PAO1 strain (these 20 bases are called spacers, and NGG is not included). The feature of this step is: in order to enable the spacer fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com