Construction of CRISPR-Cpf1-based efficient gene editing system
A gene editing and gene technology, applied in the field of high-efficiency gene editing system construction, can solve the problems that the efficiency of gene knock-in and large fragment deletion has not been verified, there is no strategy for multi-gene deletion, and the deletion efficiency needs to be improved. The effect of efficient knockout efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Construction of CRISPR-Cpf1 system and its application in single gene knockout
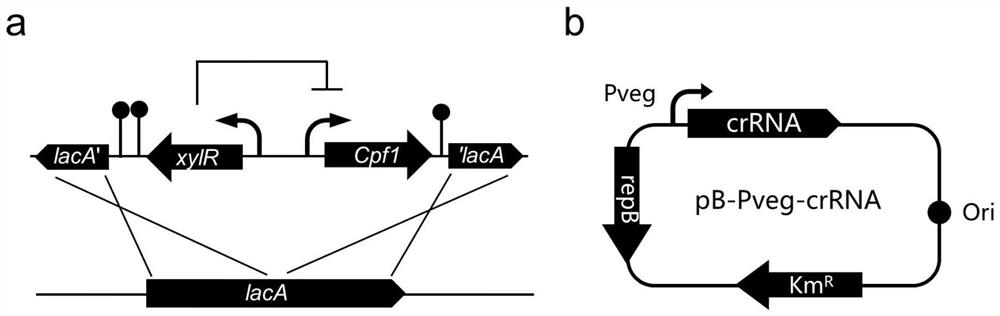
[0034] Select the Cpf1 gene (nucleotide sequence as shown in SEQ ID NO.1) from F.novicida, the Cpf1 gene is cloned to the downstream of the xylose promoter of the Bacillus subtilis integration vector pAX01 (the recombinant vector is integrated into the Bacillus subtilis 168 genome lacA site), making it regulated by xylose.
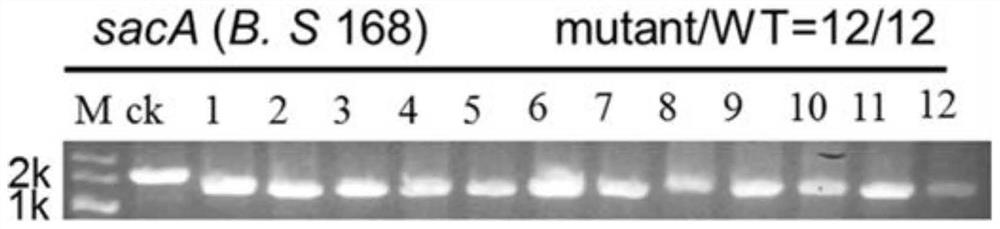
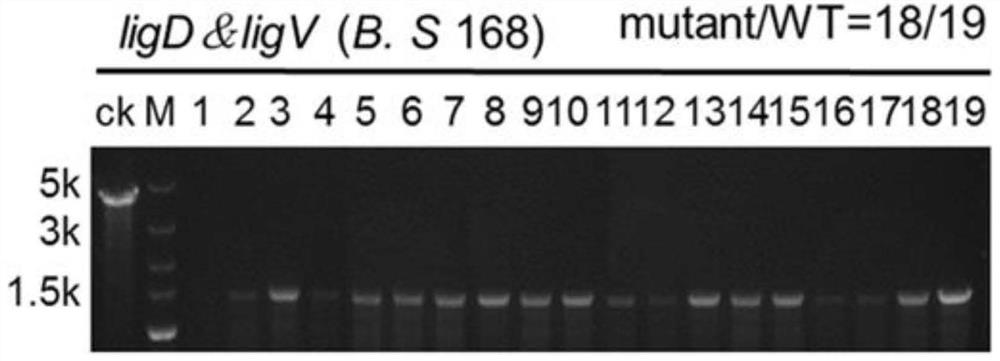
[0035]In order to fully express crRNA, the high-copy vector pB-Pveg-sfGFP (pBSG03) was selected as the expression vector of crRNA, and the crRNA targeting sacA, ligDV and pps genes were respectively cloned into pB-Pveg-sfGFP and replaced The sfGFP gene was constructed to obtain pB-Pveg-crRNA, forming pB-Pveg-sacAcrRNA, pB-Pveg-ligDVcrRNA and pB-Pveg-ppscrRNA.
[0036] Three different recombinant plasmids were transformed into Bacillus subtilis in which the Cpf1 gene was integrated at the lacA site of the genome ( figure 1 ), relevant primers are provided (s...
Embodiment 2
[0048] Example 2: Application of CRISPR-Cpf1 system in single gene knock-in
[0049] In order to reflect the knock-in efficiency of this system, the sfGFP gene was knocked into the sacA locus. The crRNA design of sacA is the same as that of sacA knockout. The sfGFP gene was connected to pB-Pveg-sacAcrRNA by seamless cloning, and the primers required for the connection were as follows (Table 3), and the recombinant plasmid pB-Pveg-sacAcrRNA-sfGFP was constructed. The recombinant Bacillus subtilis obtained by transforming the recombinant plasmid pB-Pveg-sacAcrRNA-sfGFP into the lacA site of the genome and integrating the Cpf1 gene in Bacillus subtilis was cultured in LB medium at 37°C and 200rpm at OD 600 After reaching about 0.4, xylose with a final concentration of 1% (1g / 100mL) was added to induce the expression of Cpf1 for 12 hours, and whether the target gene was knocked in was judged by colony PCR.
[0050] Through verification, the knock-in efficiency of sfGFP is as hig...
Embodiment 3
[0053] Example 3: Application of CRISPR-Cpf1 system in multiple gene knockout
[0054] Two genes, sacA and aprE, were selected as target genes and deleted at the same time. The primers for constructing crRNA are shown in Table 2. The crRNA expression cassette of aprE was cloned into pB-Pveg-sacAcrRNA to form pB-sacA-aprEKO. Then the plasmid was transformed into the Bacillus subtilis with Cpf1 integrated in the genome, and the obtained recombinant Bacillus subtilis was induced by colony PCR in LB medium at 37°C, 200rpm, and 1% xylose (1g / 100mL) for 20h. Verify the knockout efficiency of double genes. The results showed that the double gene knockout efficiency of integrated CRISPR-Cpf1 was as high as 58.3%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com