SSR fluorescence labeled primer for paternity test of pangolin and application thereof
A paternity test and fluorescent labeling technology, applied in the field of animal paternity test, can solve problems such as difficulty in individual identification and unclear pedigree, save cost and time, and protect genetic diversity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1 Screening of Malayan pangolin microsatellite markers
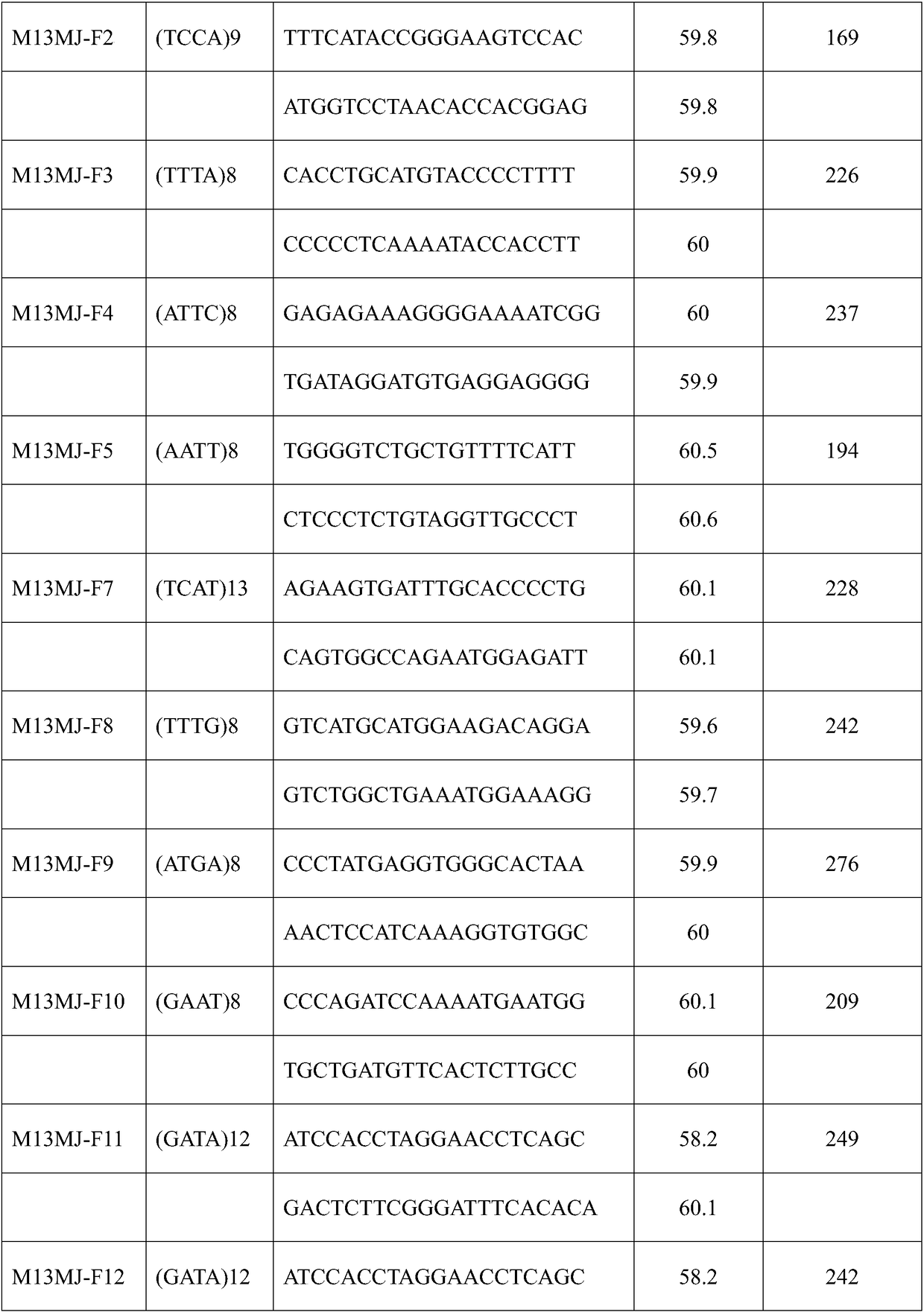
[0039] Dissected Malayan pangolin (dead) tongue, stomach, large intestine, pancreas, liver, saliva and other samples were mixed and ground, and total RNA was extracted for transcriptome sequencing, splicing, assembly and searching for microsatellite sites. A total of 18,693 SSR sites were detected, including 12,120 mononucleotide repeats, 3,202 dinucleotide repeats, 1,594 trinucleotide repeats, 259 tetranucleotide repeats, and pentanucleotide repeats. There are 52 repeat sequences and 23 hexanucleotide repeat sequences. According to the obtained SSR loci, 56079 pairs of primers were designed using Primer 5.
[0040] DNA was extracted from the saliva samples of 35 Malayan pangolins (15 females and 20 males) using the Animal Tissue DNA Mini Kit (Hipure Tissue DNA Mini Kit, Magen), and the DNA concentration and quality were detected by a NanoDrop DN-1000 UV spectrophotometer; Dilute the obtained DNA with do...
Embodiment 2
[0046] Example 2 PCR Amplification and Genotyping of Polymorphic Microsatellite Sites
[0047] (1) Microsatellite fluorescently labeled primer PCR reaction
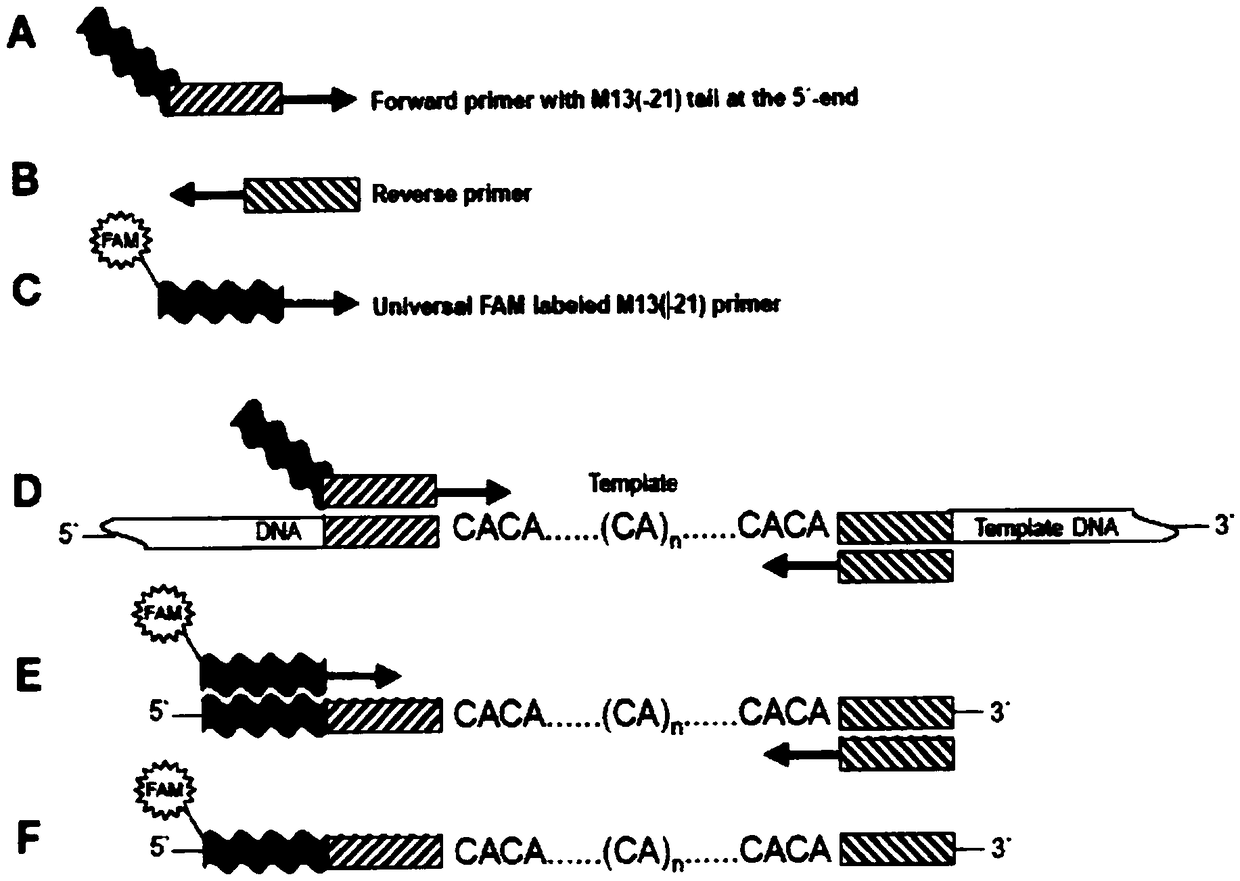
[0048] Add a FAM fluorescent group to the forward 5' end of the universal primer M13 for modification (Boutin-Ganache et al., 2001) to obtain a FAM-modified M13 forward primer. The sequence of the M13 forward primer is CACGACGTTGTAAAAC GAC; for Example 1 Preliminary analysis screened out 15 pairs of primers with polymorphic sites, and added M13 sequence (CACGACGTTGTAAAACGAC) to the forward 5' end of the primers to synthesize forward primers with M13 linker at the 5' end.
[0049] The saliva sample DNA of 35 Malayan pangolins extracted in Example 1 was used as a template for fluorescent PCR reaction. The amplification system was 10 μl, including 1 μl of 100ng / μl DNA template, 5 μl of 2×PCR Mix, and 1.6 μl of ddH2O, which had polymorphism 0.4 μl of forward primer with M13 linker added to the forward 5’ end of the 15 pairs ...
Embodiment 3
[0059] Example 3 Application of Microsatellite Markers in Malayan Pangolin Paternity Test
[0060] The above-mentioned 15 microsatellite markers were used in conjunction with the exclusion method and the likelihood method to identify the paternity of 35 Malayan pangolins, and compared the results of the personal identification with the pedigree records of the farm to verify the accuracy of the method. The LOD value calculated by using CERVUS 3.0 software is 5.75 at the 95% confidence level and 2 at the 80% confidence level. When the confidence level is 95%, it is identified that number 246 and C46, A26 and BB are mother-child relationship; A25 and BB are identified as father-son relationship. Comparing the personal identification results with the pedigree records gave a 100% accuracy rate.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com