Kit for detecting drug-resistant mutation in YMDD motif region of hepatitis B virus
A hepatitis B virus and drug-resistant mutation technology, which is applied in the direction of recombinant DNA technology, microbial measurement/testing, DNA/RNA fragments, etc., can solve the detection of drug-resistant mutations in the hepatitis B virus YMDD motif region that has not been queried, etc. problem, achieve the effect of shortening the detection time, reducing the possibility of pollution, and improving efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
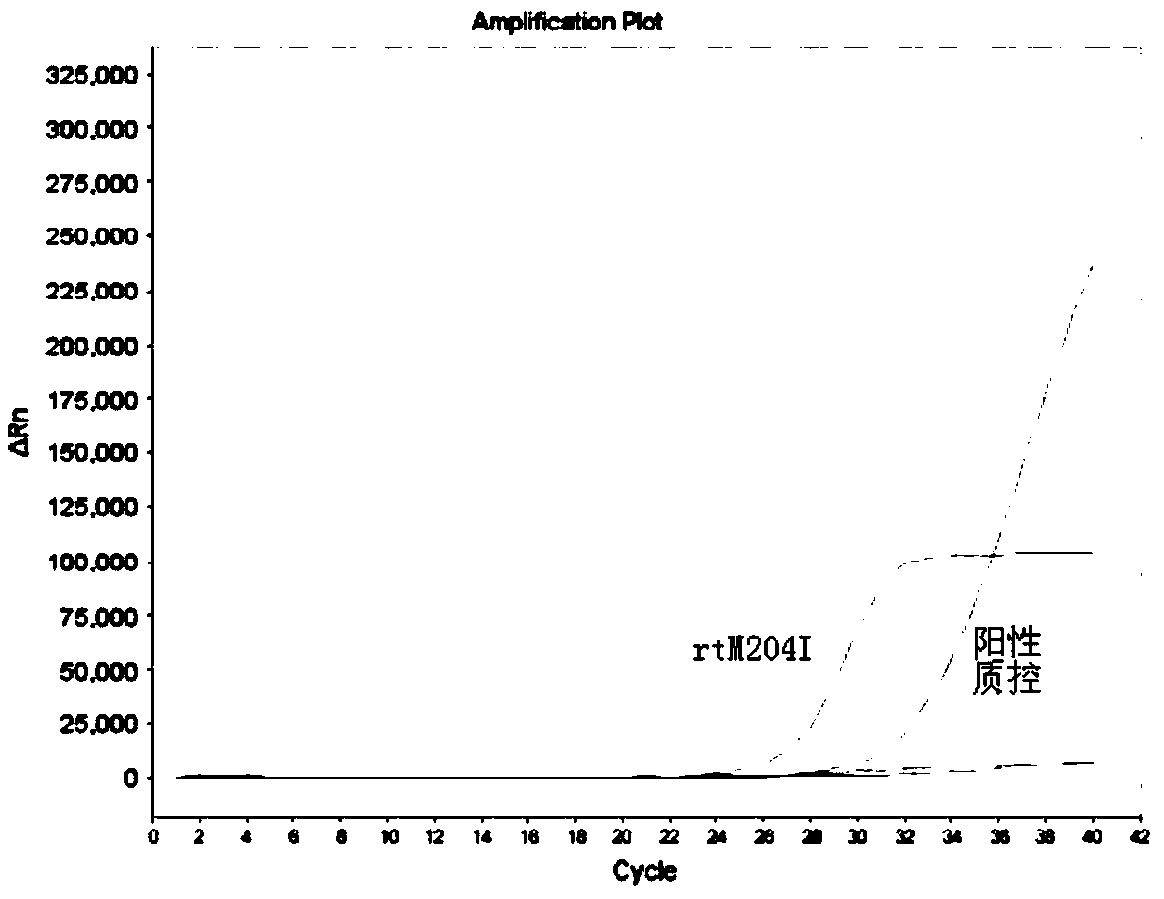
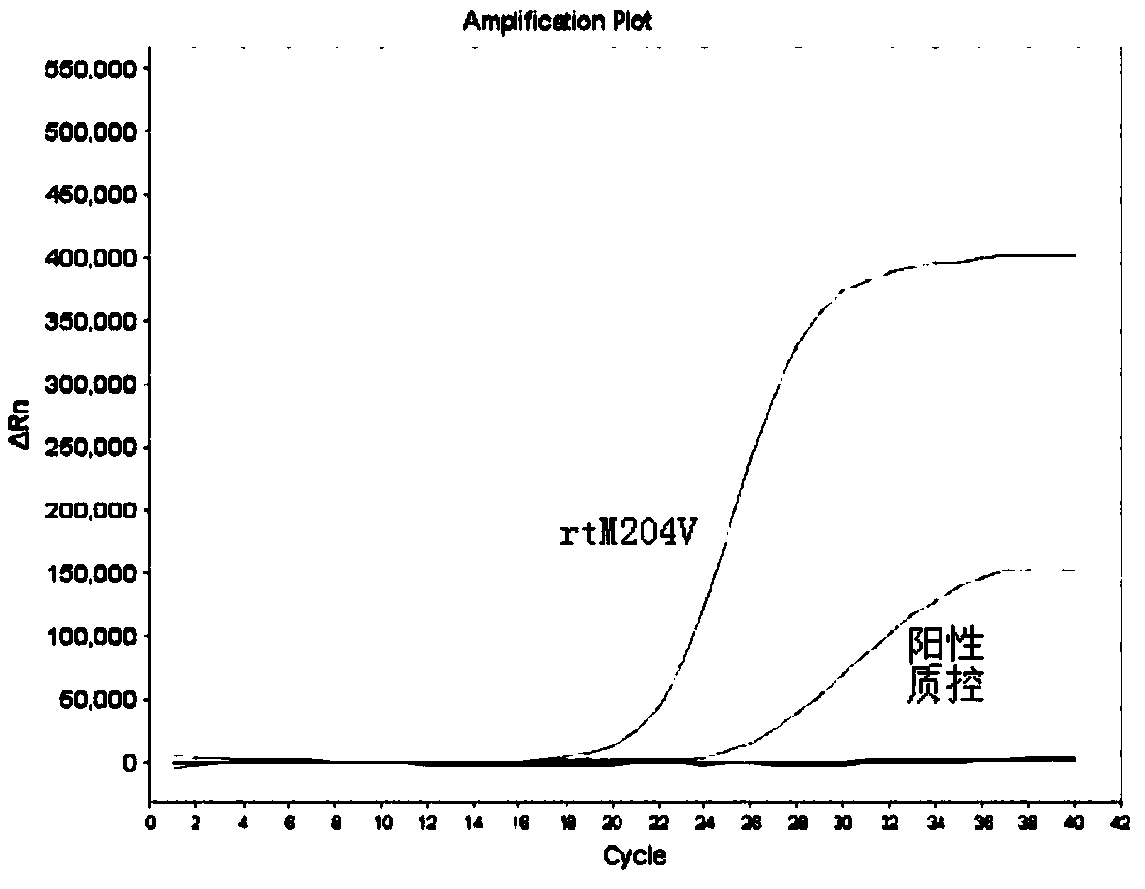
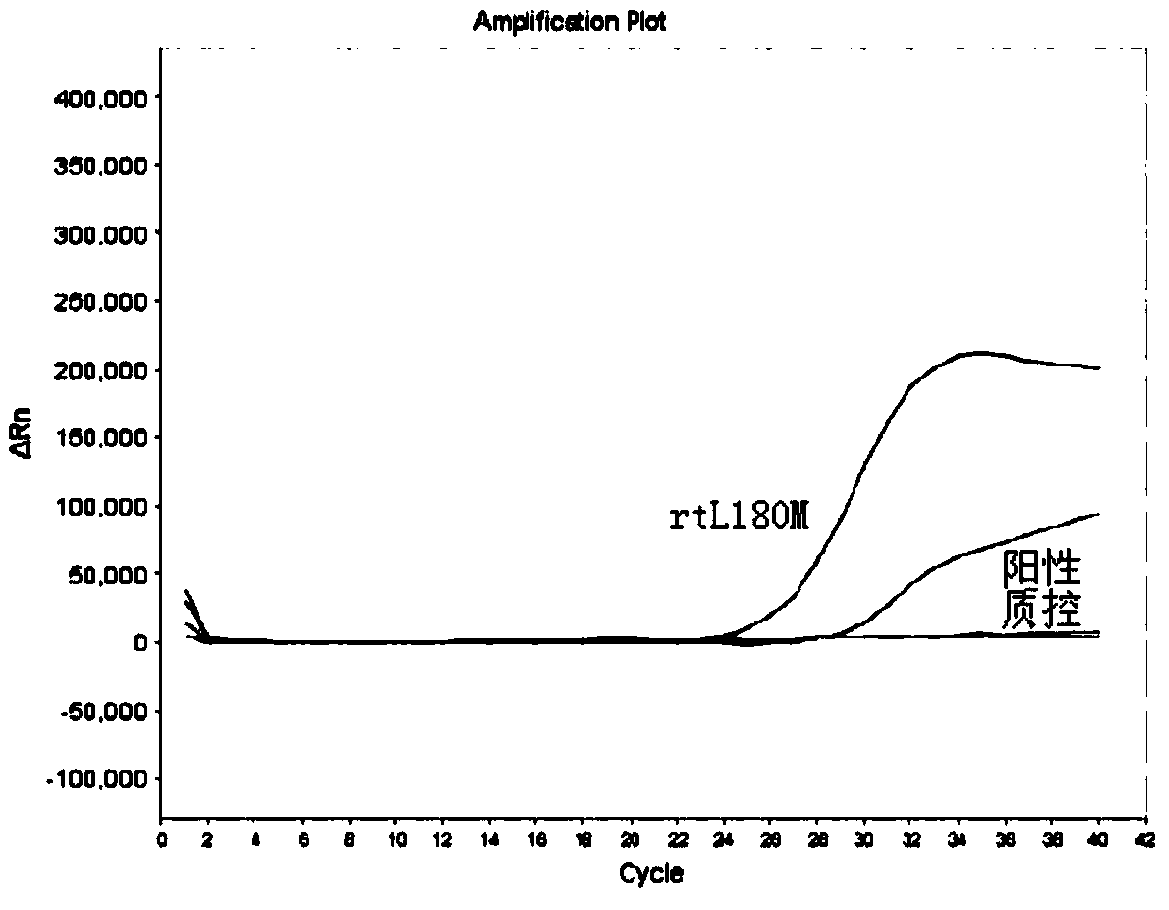
[0033] The present embodiment has designed the primer probe directed at the drug-resistant mutation of hepatitis B virus YMDD motif region, designed the specific primers SEQ ID NO.1 and SEQ ID NO.2 directed at the rtM204I mutation site, and directed at the rtM204V mutation site. Specific primers SEQ ID NO.1 and SEQ ID NO.3, and the probe SEQ ID NO.6 corresponding to the rtM204I / V mutation site; specific primers SEQ ID NO.4 and SEQ ID NO for the rtL180M mutation site .5, and the probe SEQ ID NO.7 corresponding to the rtL180M mutation site. The specific sequence is as follows:
[0034] 5'-AGTCCGTTTCTCTTGGCTC-3, the sequence is shown in SEQ ID NO.1;
[0035] 5'-CCCCAAWACCACATCATCaAAATA-x, the sequence is shown in SEQ ID NO.2;
[0036] 5'-CCAAWACCACATCATCCATCCAcATAAC-x, the sequence is shown in SEQ ID NO.3;
[0037] 5'-GGCCTCAGTCCGTTTCTCaTGG-x, the sequence is shown in SEQ ID NO.4;
[0038] 5'-AGCGGCATAAAGGGACTC-3', the sequence is shown in SEQ ID NO.5;
[0039] FAM-GGGAAAGCC...
Embodiment 2
[0043] This example prepares a kit for drug-resistant mutations in the YMDD motif region of hepatitis B virus.
[0044] The specific design of the kit is as follows:
[0045] (1) DNA extraction solution, including concentrated solution and lysis buffer;
[0046] (2) PCR reaction master mix 1: the main components are PCR buffer, 4 kinds of dNTPs, probe SEQ ID NO.6, and primers mixed with SEQ ID NO.1 and SEQ ID NO.2; among them, the probe SEQ ID NO.6 The concentration of ID NO.6 is 0.25 μM, and the concentration of primers SEQ ID NO.1 and SEQ ID NO.2 is 0.25 μM.
[0047] (3) PCR reaction master mix 2: the main components are PCR buffer, 4 kinds of dNTPs, probe SEQ ID NO.6, and primers mixed with SEQ ID NO.1 and SEQ ID NO.3; among them, the probe SEQ ID NO.6 The concentration of ID NO.6 is 0.25 μM, and the concentration of primers SEQ ID NO.1 and SEQ ID NO.3 is 0.25 μM.
[0048] (5) PCR reaction master mix 3: the main components are PCR buffer, 4 kinds of dNTPs, probe SEQ ID N...
Embodiment 3
[0054] In this example, the kit for detecting drug-resistant mutations in the YMDD motif region of hepatitis B virus prepared in Example 2 is used to detect samples, and the specific detection steps are as follows.
[0055] (1) Extracting the HBV DNA template: the HBV DNA template in the serum sample of hepatitis B virus was extracted with DNA extraction solution.
[0056] The specific process includes: adding 100 μL of concentrated solution to hepatitis B virus serum, centrifuging at 12000 rpm for 10 minutes; discarding the supernatant; adding 25 μL of lysis buffer and mixing, lysing at 100 °C for 10 minutes; finally centrifuging at 12000 rpm for 10 minutes, and the supernatant is ready for use HBVDNA template.
[0057] (2) PCR amplification detection: use the HBV DNA extracted in step (1) as a template, and use the kit provided by the present invention to perform corresponding amplification detection.
[0058] Mix PCR reaction premixes 1, 2 and 3 with the enzyme mixture in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com