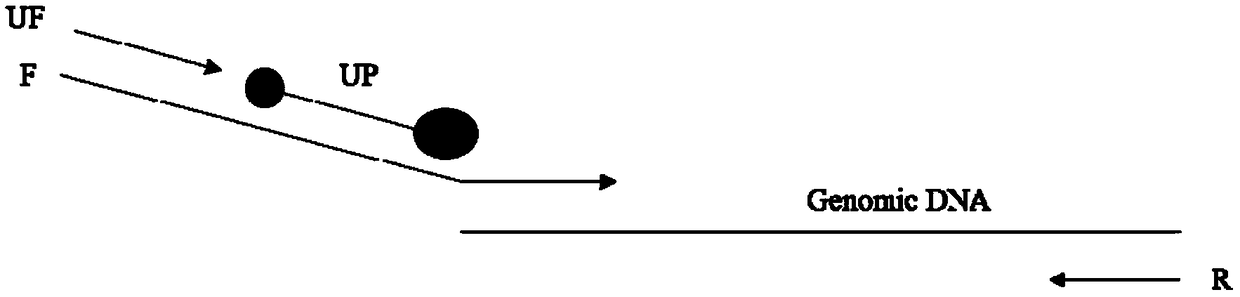
Rare mutation detection method, kit and application thereof
A detection kit and a technology for rare mutations, applied in the field of molecular biology, can solve the problems of different mismatch discrimination capabilities, only 20% sensitivity, and easy contamination of non-closed tubes, to achieve enhanced mutation base recognition capabilities, high sensitivity, The effect of reducing pollution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] According to the missense mutation at position 2369 of the EGFR gene, that is, the position 2369 is changed from C to T (T790M for short), the wild-type plasmid and the mutant-type plasmid were respectively constructed as templates through genetic engineering. The sequence of the wild-type plasmid is shown in SEQ ID No: 1. After determining the concentration of the plasmid, dilute it to 1ng / ul with TE buffer, and then carry out a 10-fold gradient dilution of the 1ng / ul plasmid with TE buffer, and the dilution concentration is 10 -1 、10 -2 、10 -3 、10 -4 、10 -5 、10 -6 ng / ul spare, 10 -6 The gene copy number contained in ng plasmid is equivalent to the target gene copy number contained in 1ng genomic DNA.
[0068] The diluted mutant plasmid and wild-type plasmid were mixed in different proportions to obtain wild-type samples (without mutant plasmids), 1% mutant samples (the ratio of mutant plasmids to wild-type plasmids was 1:99), 5% Mutant samples (ratio of mutant p...
Embodiment 2
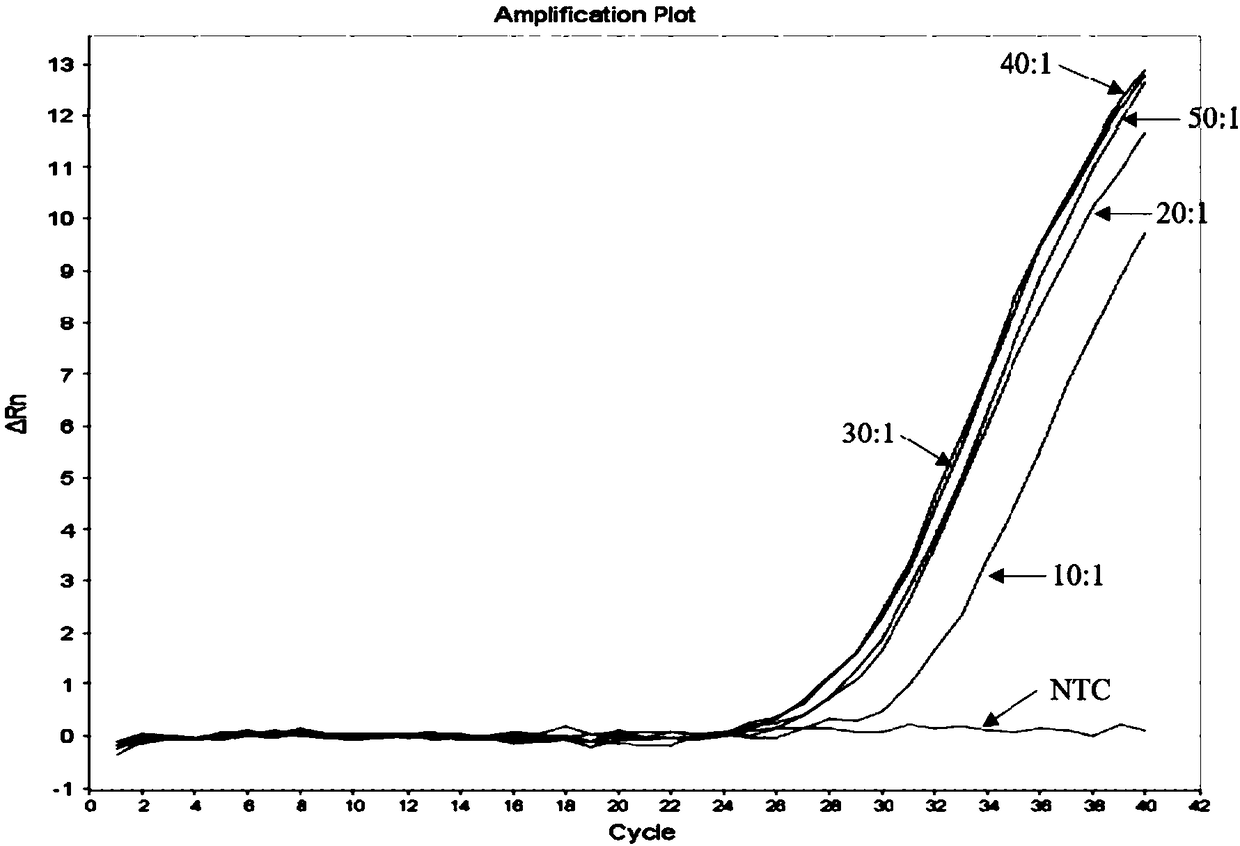
[0074] Using the mutant plasmid as a template, the nucleotide sequences of SEQ ID No:6 and SEQ ID No:2 are ARMS primers, the nucleotides shown in SEQ ID No:7 are universal primers and the nucleotides shown in SEQ ID No:8 Fluorescent PCR was carried out for the Taqman-MGB probe. In the fluorescent PCR reaction, the ratios of universal primers and AMRS upstream primers were 10:1, 20:1, 30:1, 40:1, 50:1, and the concentrations of the remaining components were the same ( The final concentration of each component in the 20μl system is 1×PCR Buffer Mix, 500nM ARMS downstream primer, 500nM universal primer, 250nM Taqman-MGB probe, 10 -6 ng mutant plasmid and balance water). Fluorescence PCR reaction condition is identical with embodiment 1, and result is as follows image 3 shown. Depend on image 3 It can be seen that when the ratio of universal primers to ARMS upstream primers is 40:1, the amplification efficiency is the best.
Embodiment 3
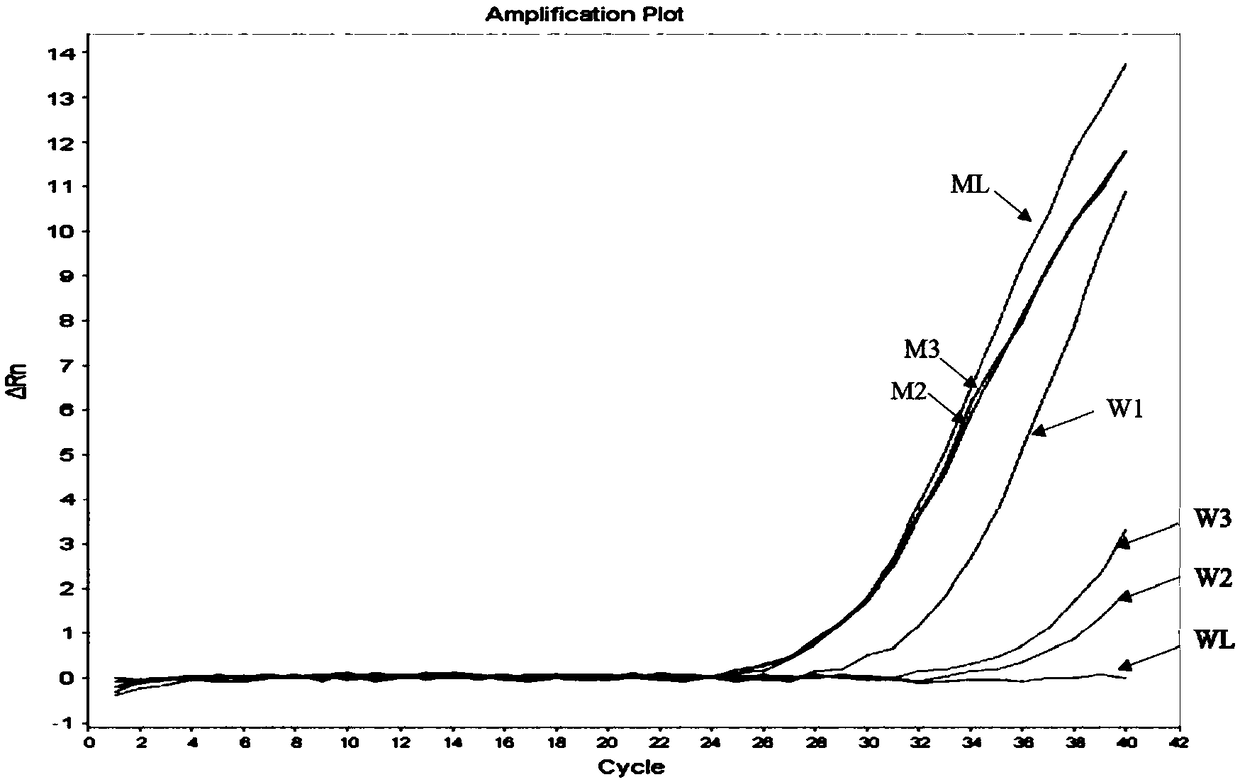
[0076] Adopt T790M site to carry out experiment, with the 100% mutant sample prepared in embodiment 1, 50% mutant sample, 20% mutant sample, 10% mutant sample, 5% mutant sample, 1% mutant sample and wild-type sample (adding amount is average for 10 -6 ng) as a template, using primers and primer ratios and Taqman-MGB probes in Example 2 to carry out fluorescent PCR to verify its sensitivity, the results are as follows Figure 4 shown. Depend on Figure 4 It can be seen that the detection sensitivity of the detection method disclosed in the present invention can reach 1%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com