Method for rapidly constructing target pU6-sgRNA plasmids required for caenorhabditis elegans gene editing
A pu6-sgRNA, Caenorhabditis elegans technology, applied in DNA/RNA fragments, recombinant DNA technology, introduction of foreign genetic material using vectors, etc. The effect of low cost, reduced working time and wide adaptability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Construction of pU6-rho-1sgRNA plasmid by traditional two-step PCR method
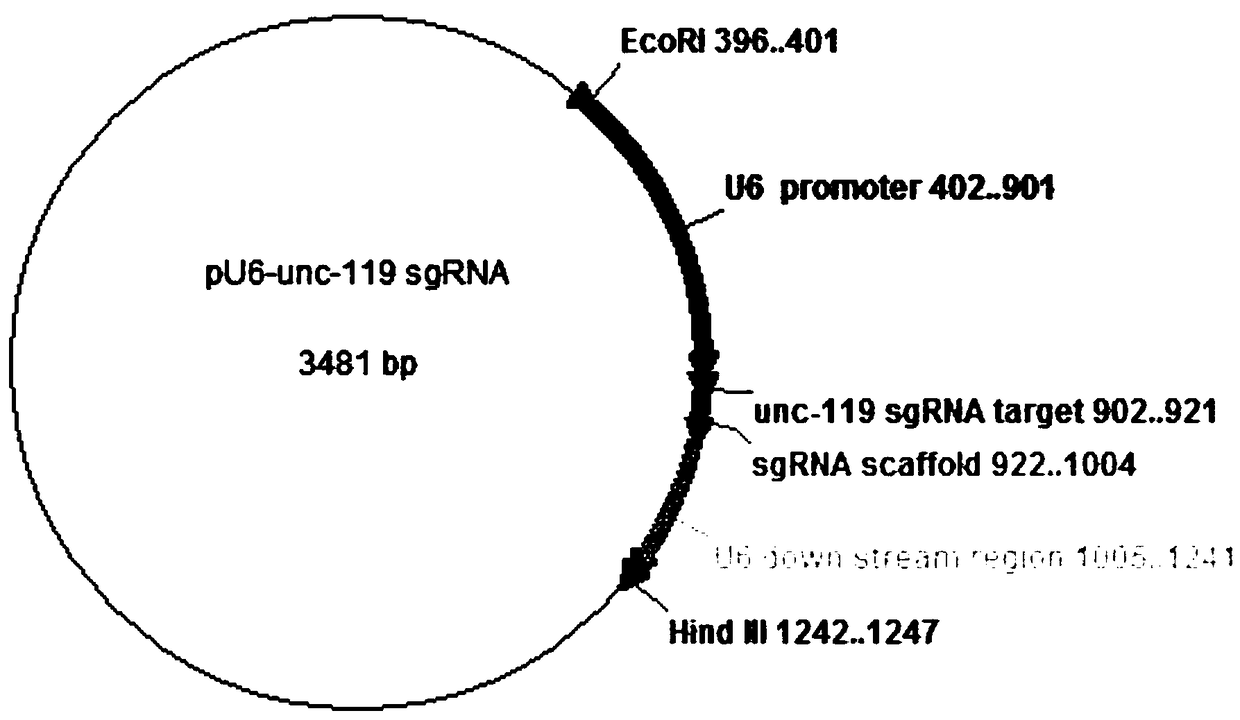
[0058] see figure 1 and figure 2 , This embodiment provides a method for constructing pU6-rho-1 sgRNA plasmid in the traditional two-step PCR method described in the background art. The technical principle is that the pU6-unc-119 sgRNA plasmid (#46169) is used as the original vector, and the target sequence of the sgRNA plasmid is shown in SEQ ID NO.2. Using this plasmid as the original vector, it is necessary to replace the 20bp sgRNA target DNA sequence of unc-119sgRNAtarget 902..921 gene loci with sgRNA target DNA sequences of different gene loci to obtain sgRNA plasmids containing different target DNA sequences.
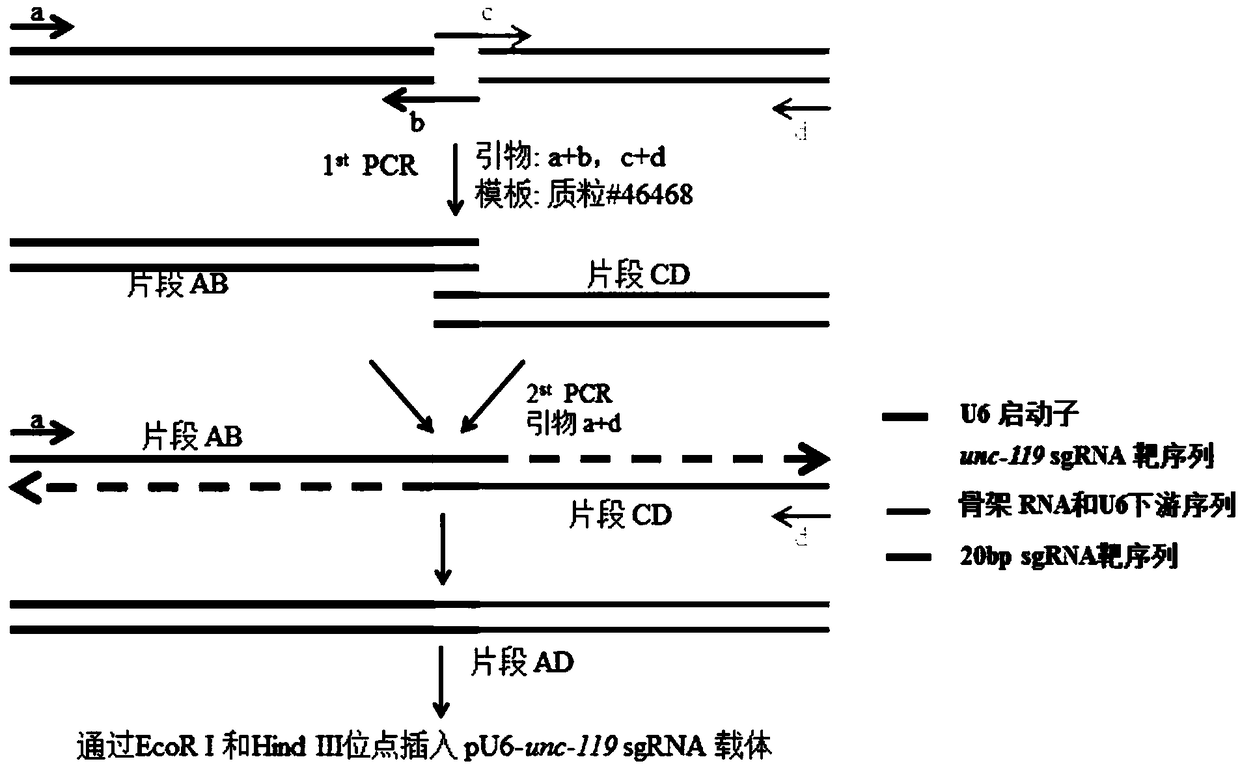
[0059] see figure 2 , using a two-step PCR method to construct pU6-rho-1sgRNA, the specific steps are as follows:
[0060] 1) Using the pU6-unc-119sgRNA plasmid as a template, digest with two restriction enzymes Eco RI and Hind III to release a 846bp fragment, including the U...
Embodiment 2
[0071] This embodiment provides an improved method for constructing pU6-rho-1sgRNA plasmid
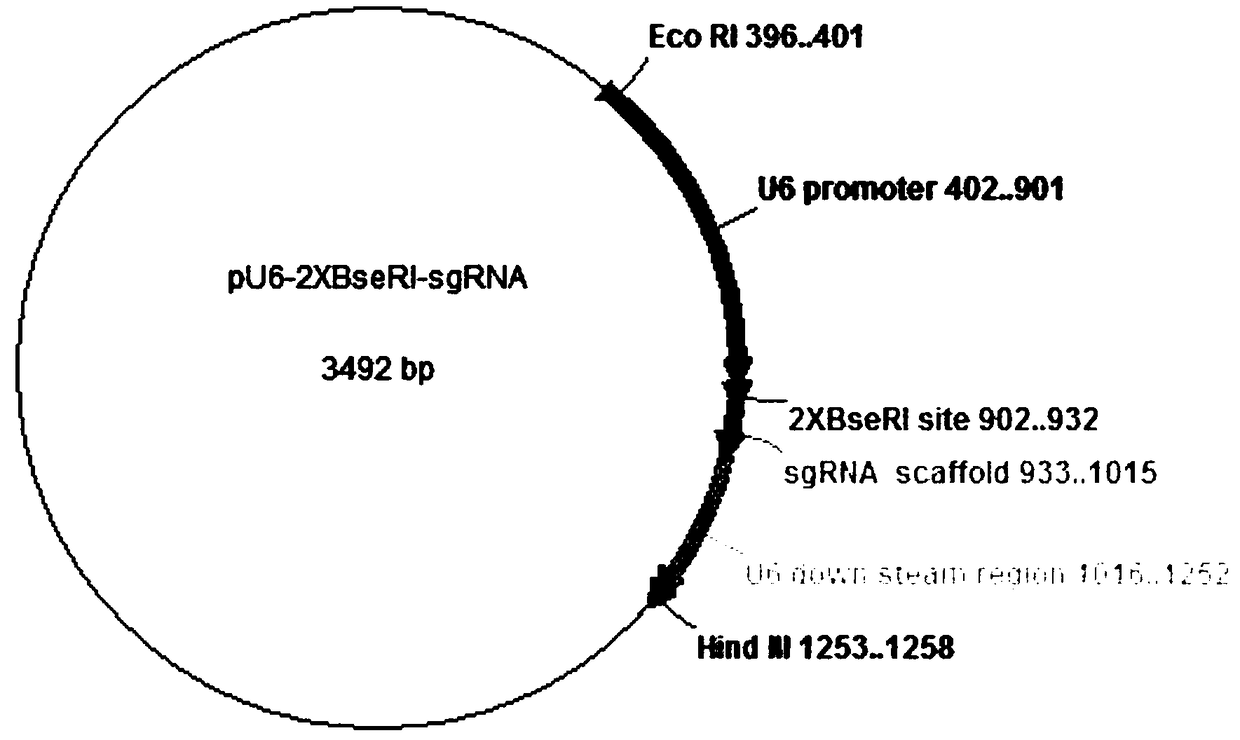
[0072] see Figure 3-Figure 5 , the inventors first transformed the original vector of pU6-unc-119sgRNA. Insert two opposite BseRI restriction sites AGCTGAGT between the U6 promoter and the sgRNA backbone sequence CTCCTC GAT GAGGAG CAGATCAG (the underlined part) and the spacer sequence are used to construct the pU6-2XBseRI-sgRNA plasmid. The subsequent plasmid construction is based on the pU6-2XBseRI-sgRNA plasmid. The specific steps for subsequent construction of the plasmid are as follows:
[0073] 1) Digestion of pU6-2X BseRI-sgRNA plasmid with BseRI: Mix 4 μL of pU6-2XBseRI-sgRNA plasmid, 10 μL of 10xCutsmart digestion buffer required for BseRI digestion, 2 μL of BseRI enzyme, and 84 μL of double distilled water in a PCR tube. Enzyme digestion at ℃ for 1h, heat shock treatment at 80℃ to terminate the digestion reaction, run 1% gel electrophoresis on the digested product, recov...
Embodiment 3
[0106] This example provides a method for constructing pU6-F33A8.4-sgRNA1 plasmid
[0107] If you need to construct other sgRNA plasmids, the relevant primers you need are:
[0108] F: 5'-N20GT-3', wherein N20 represents the first 20bp sgRNA target site sequence of PAM (NGG);
[0109] R: 5'-n20AA-3', wherein n20 represents the reverse complementary sequence of the first 20 bp sgRNA target site sequence of PAM (NGG);
[0110] The difference from Example 2 is that the target site sequence and the primer sequence to be synthesized are:
[0111] The F33A8.4-sg1 target site sequence is GAAGGACCATGAGAGCACTG (as shown in SEQ ID NO.12);
[0112] Primers to be synthesized:
[0113] F33A8.4-sg1-F: GAAGGACCATGAGAGCACTGGT (as shown in SEQ ID NO.13);
[0114] F33A8.4-sg1-R: CAGTGCTCTCATGGTCCTTCAA (as shown in SEQ ID NO.14);
[0115] The gel electrophoresis images of screened positive bacterial liquid clones are as follows: Figure 8 As shown, the positive rate of pU6-F33A8.4-sgRNA1 p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com