A method for detection of susceptibility gene of congenital edentulousness
A susceptibility gene and congenital technology, applied in the field of gene chips, can solve the problems of large amount of sequencing results, high price, time-consuming, etc., and achieve the effect of enriching the spectrum of pathogenic mutations, expanding the coverage of diagnosis, and facilitating the interaction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
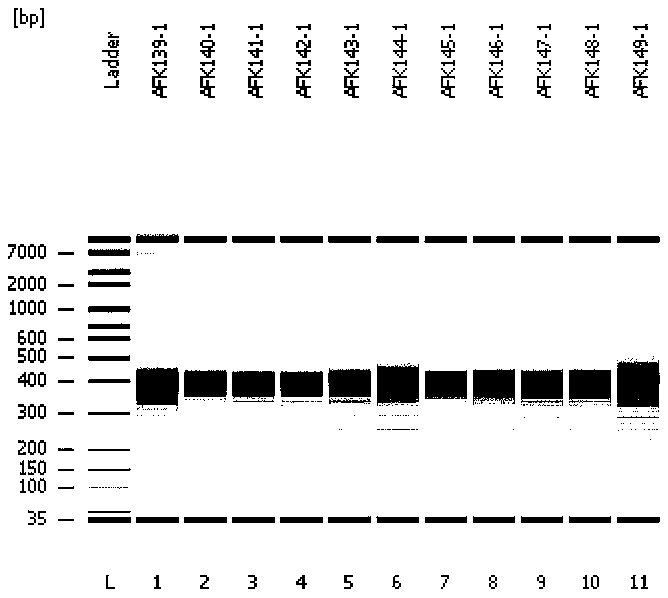
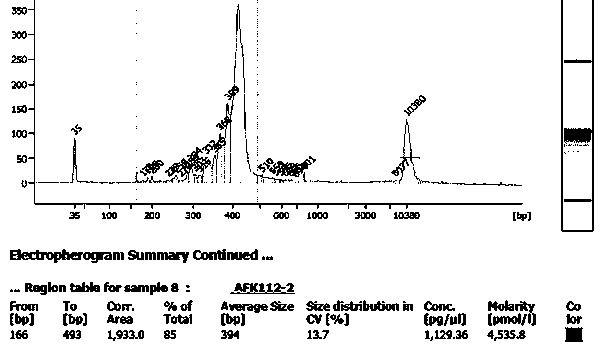
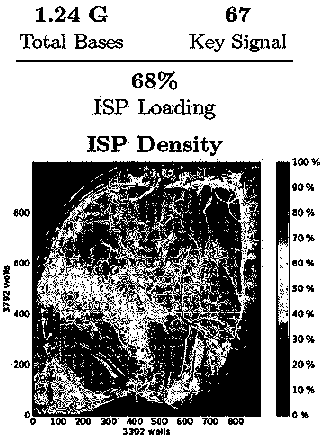
Image
Examples
Embodiment 1
[0027] Example 1 Acquisition of DNA samples to be sequenced
[0028] 1) Collect 2 mL of saliva samples from patients, family members and normal controls, and collect them in the saliva sample collection tubes in the Oragene DNA (OG-500) DNA collection kit (DNA Genoteck, general agent of Bipi Biotechnology Co., Ltd.), at room temperature Save it.
[0029] 2) Use the Oragene·DNA (OG-500) DNA collection kit (DNA Genoteck, the general agent of Bipi Biotechnology Co., Ltd.) to extract human genomic DNA from the collected saliva samples.
[0030] 3) 1 μl of the extracted DNA was loaded as a sample, and the amount of genomic DNA was checked by 1% agarose gel electrophoresis, and the DNA was quantified with a UV-640 Beckman spectrophotometer. Aliquot with appropriate volume and freeze at -20°C.
Embodiment 2
[0031] The screening of embodiment 2 congenital edentulous gene
[0032] Collect the reported genes that may be related to congenital edentulousness, according to the importance of gene expression in the development of tooth germ, the frequency of occurrence in various literatures or reports, data and other factors, from more than 60 congenital edentulous 35 edentulous candidate genes were screened out, and classified according to whether mutations were detected in human congenital edentulous patients, as shown in Table 1:
[0033] Table 1: Selected congenital edentulous genes
[0034]
[0035]
[0036] The test samples used in the detection method of congenital edentulous susceptibility gene of the present invention come from patients, family members and normal controls, and can detect the gene mutations in the 35 gene sequences in Table 1 of congenital edentulous patients On the one hand, use clinical big data to verify the existing research and development results, a...
Embodiment 3
[0038] 1. Reagents for detection
[0039] Library Construction Reagents:
[0040] (1) Primer library (synthesized by Life Technologie)
[0041] (2) Ion AmpliSeq TM Library Kits 2.0(Life Technologies)
[0042] (3)Ion Xpress TM Barcode Adapters (Life Technologies)
[0043] (4) XP Reagent (Beckman Coulter)
[0044](5) Agilent High Sensitivity DNA Kit (Agilent)
[0045] Ion OneTouch TM Reagent: Ion PGM TM Hi-Q TM OT2Kit (Life Technologies)
[0046] Ion Torrent sequencing reagent: Ion PGM TM Hi-Q TM Sequencing Kit (Life Technologies)
[0047] 2. Experimental process
[0048] (1a) Use two sets of primer libraries (see Example 7 for the sequences of the primer libraries) to amplify the full-length gene in the target region to obtain preliminary sequencing library fragments. The amplification system and PCR amplification conditions are as follows:
[0049] Table 2: Amplification System
[0050] Element Amount added AmpliSeq TM HiFi Mix(red cap)
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com