Identification and application of SNP in the fourth exon region of porcine incenp
A technique for boars and large white pigs, applied in DNA/RNA fragmentation, recombinant DNA technology, determination/inspection of microorganisms, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Method used
Image
Examples
Embodiment 1
[0021] Example 1 Extraction of Boar Semen Genomic DNA Using Phenol Extraction
[0022] Inject the fresh semen of the boar collected on the spot (from the Yaji Mountain Pig Artificial Insemination Center of Guangxi Yangxiang Pig Gene Technology Co., Ltd., the American Duroc, Landrace and Large White breeds, the same below) into 10ml centrifuge tubes and stored frozen at -20°C. Transported to the laboratory below 0°C, then thawed in a water bath, and extracted boar semen genomic DNA. Specific steps are as follows:
[0023] (1) Take 1ml of pig semen, put it in a 2ml centrifuge tube, centrifuge at 5000rpm for 7min, discard the supernatant.
[0024] (2) Add 1000ul sperm washing solution (1M Tris-Hcl (PH=8.0) 10ml, 0.5M EDTA (PH=8.0) 20ml, 5M NaCl 100ml to each centrifuge tube, add ddH 2 (870ml, mix well, conventional high-pressure steam sterilization, store at room temperature) or normal saline, blow and beat repeatedly, centrifuge at 12000rpm for 7min, and discard the supernata...
Embodiment 2
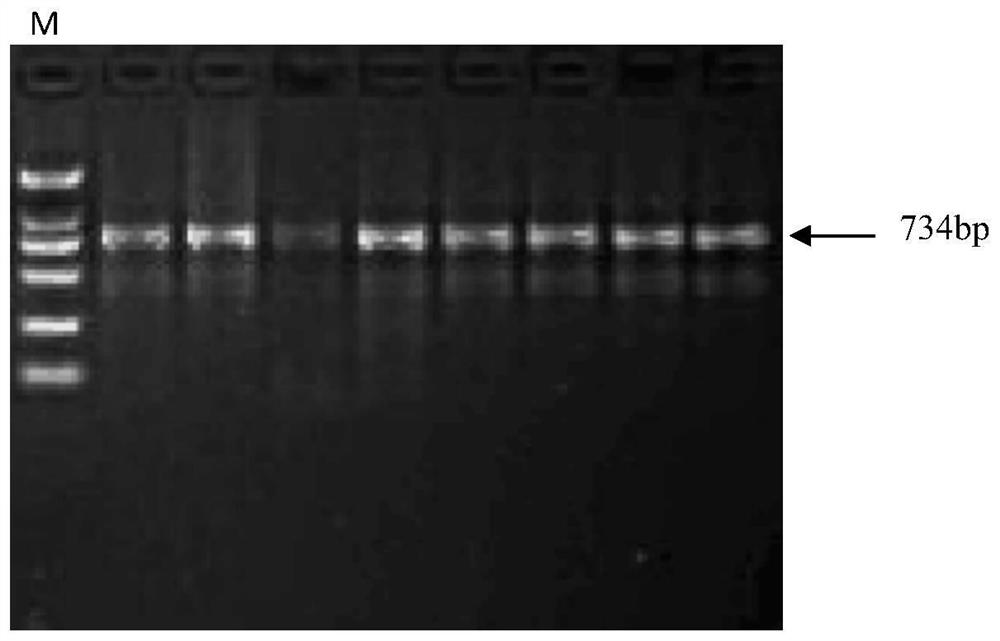
[0033] Embodiment 2: the acquisition of boar INCENP fourth exon region-specific DNA fragment and the establishment of single nucleotide polymorphism (SNP) detection method
[0034] Select pigs of foreign blood, such as American "Duroc", "Landrace", and "Large White" (from the Yaji Mountain Pig Artificial Insemination Center, a subsidiary of Guangxi Yangxiang Pig Gene Technology Co., Ltd., which are routinely reported American breeds , the implementation of the present invention is not limited to the above-mentioned varieties) take the mRNA sequence of pig INCENP gene (GeneBank accession number is NC_010444) as seed, compare in the genome of pig, design following primers according to its fourth exon sequence:
[0035] Forward primer F: 5'CTCCGCAGCAAGGACAAGG 3',
[0036] Reverse primer R: 5'AGGCACAAAGGACCACCCAC 3';
[0037] Taking Duroc pig, Landrace pig, and Large White pig blood genome DNA as templates respectively, carry out PCR amplification with the above-mentioned primer ...
Embodiment 3
[0047] Example 3: Association analysis and application of molecular markers cloned in the present invention and reproductive traits of boars
[0048] Statistics of Genotype Frequency and Allele Frequency
[0049] Genotype frequency: refers to the proportion of a specific genotype in a population to all genotypes in the population. The statistical method of genotype frequency is: genotype frequency = number of individuals of this genotype / total number of measured populations
[0050] Allele frequency: refers to the relative proportion of a gene in a population to the total genes at that locus. Statistical method of allele frequency: genotype frequency of homozygote of this gene + genotype frequency of heterozygote containing this gene / 2
[0051] Table 3 The genotype frequency and allele frequency of the SNP in the fourth exon region of INCENP in two breeds of pigs
[0052]
[0053] In order to determine whether the SNP in the fourth exon region of the pig INCENP gene is r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com