CD4 helper T cell epitope fusion peptide and vaccines thereof
A fusion peptide and auxiliary technology, applied in the fields of molecular biology and immunology, can solve the problems of low level of cellular immune response, inability to meet the needs of tumor vaccines, difficulty in playing auxiliary functions, etc., and achieve the effect of effectively enhancing efficacy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1 Construction of DNA vaccine pVKD1.0-hLMN
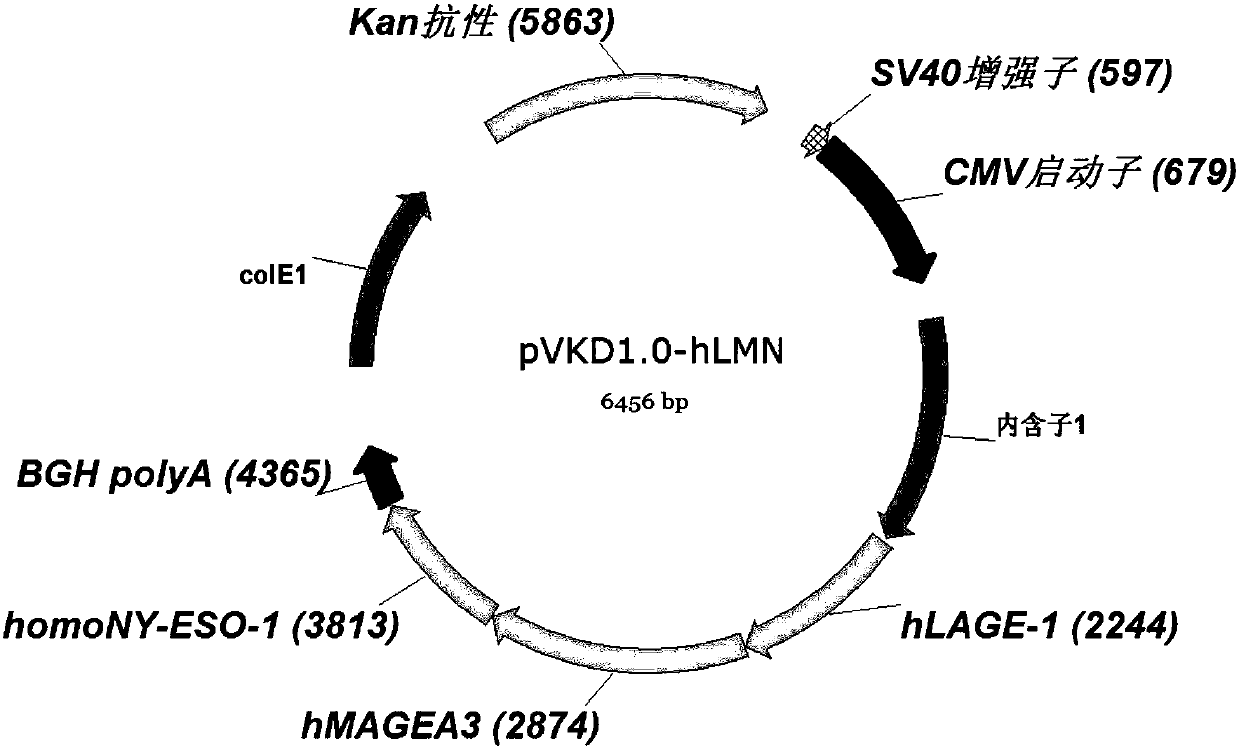
[0054] The amino acid sequences of LAGE-1, MAGE-A3 and NY-ESO-1 are shown in SEQ ID NO: 24-26, respectively. The amino acid sequence of the above antigen was optimized into the nucleotide sequence of mammalian codon usage preference by online codon optimization software (http: / / www.jcat.de / ), as shown in SEQ ID NO:27-29 respectively. After being synthesized by Shanghai Jierui Biotechnology Co., Ltd., it was cloned into the multiple cloning site between Sal I and BamH I on the DNA vaccine vector pVKD1.0 (provided by Suzhou Industrial Park Weida Biotechnology Co., Ltd.) In between, a DNA vaccine vector pVKD1.0-hLMN (plasmid map as shown in figure 1 ), which were correctly identified by sequencing and entered into the library. The vector pVKD1.0-hLMN was identified with restriction endonucleases Sal I and BamHI (enzyme digestion system is shown in Table 1), and its enzyme digestion verification map is as follows fi...
Embodiment 2
[0057] Example 2 Construction of DNA vaccine pVKD1.0-hLMN-CTB
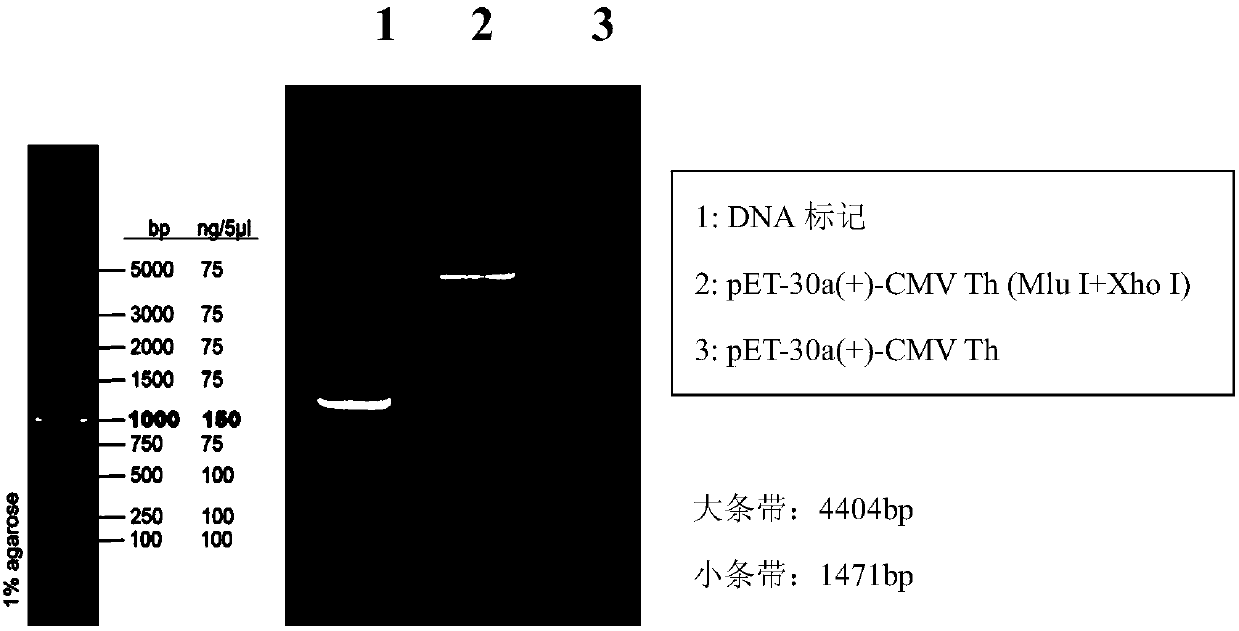
[0058] The mammalian codon-optimized sequence (SEQ ID NO: 31) of the amino acid sequence (SEQ ID NO: 30) of Cholera toxin subunit B (CTB) and its eukaryotic expression vector pVKD1.0-CTB were provided by Suzhou Provided by Weida Biotechnology Co., Ltd. in the industrial park. Using pVKD1.0-CTB as a template, design primers (Table 2), amplify the CTB gene fragment by PCR, then recover the corresponding fragment from the gel, insert the CTB fragment into the corresponding position of the linearized vector pVKD1.0-hLMN by homologous recombination Above, construct the DNA vaccine vector pVKD1.0-hLMN-CTB (plasmid map as image 3 ), which were correctly identified by sequencing and entered into the library. The vector pVKD1.0-hLMN-CTB was identified with restriction endonucleases Sal I and BamH I (enzyme digestion system is shown in Table 3), and its digestion verification map is as follows Figure 4 shown.
[005...
Embodiment 3
[0063] Example 3 Construction of DNA vaccine pVKD1.0-CI-LMNB
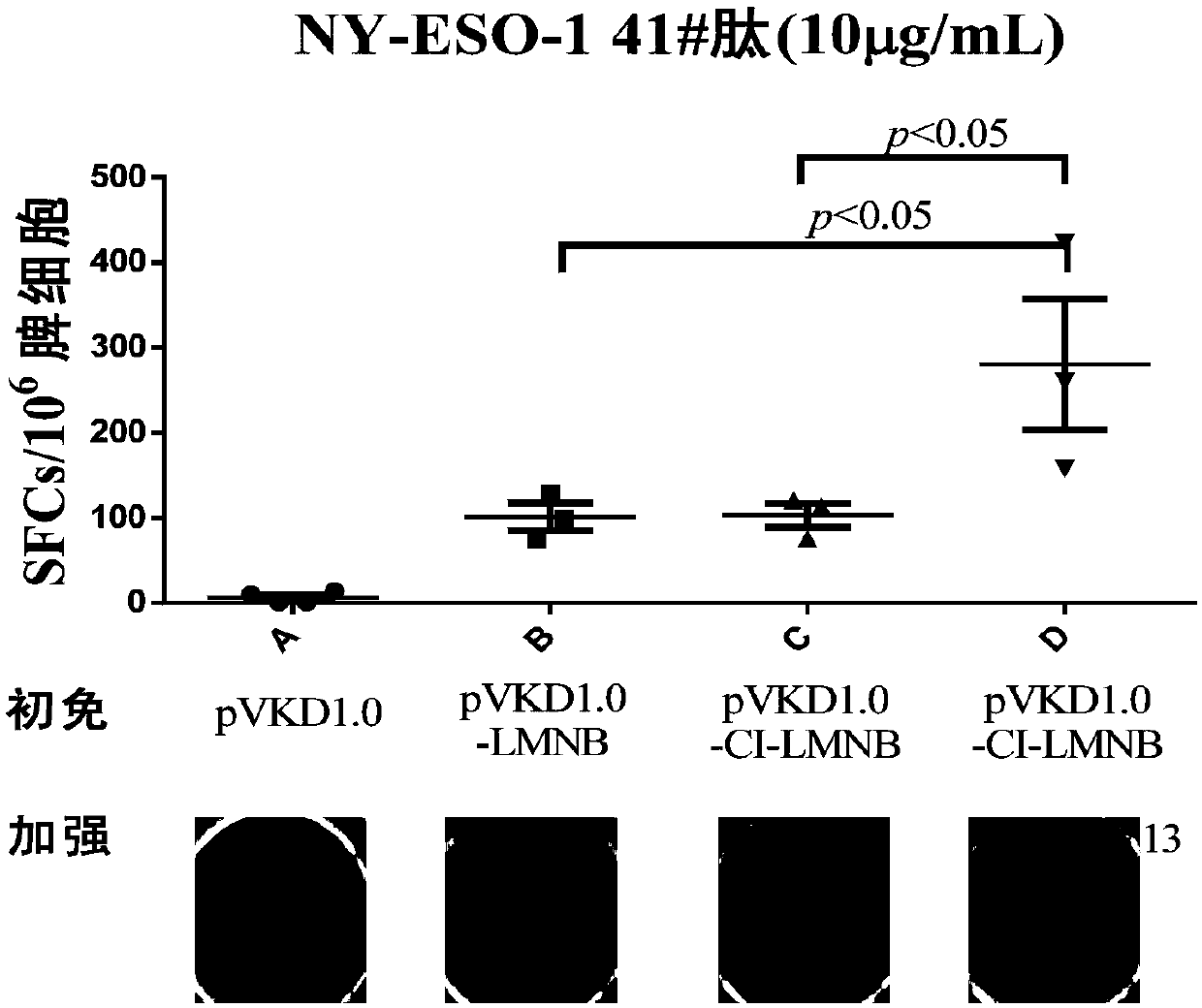
[0064] Strong Th epitopes (see Table 4) derived from cytomegalovirus (Cytomegalovirus, CMV) and influenza virus (Influvirus, Flu) were obtained on the immune epitope database (IEDB, http: / / www.iedb.org), wherein , strong Th epitopes of CMV include pp65-11, pp65-71, pp65-92, pp65-123, pp65-128, pp65-57, pp65-62, pp65-30, pp65-112, and pp65-104; influenza virus Strong Th epitopes include HA203, NP438, NS1-84, M1-181, HA375, NP24, NP95, NP221, HA434, HA440, NP324, M1-127, and M1-210. The epitopes selected in Table 4 cover most subtypes of MHC class II molecules in the human population, and also cover the subtypes of MHC class II molecules in mice. Then the selected epitopes pp65-11, pp65-71, pp65-92, pp65-123, pp65-128, HA203, NP438, NS1-84, M1-181, HA375, NP24, NP95, NP221 are concatenated together, An epitope fusion peptide of CMV virus and influenza virus is formed, the amino acid sequence of which is shown in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com