Improved method for selecting polypeptide producing cells
A technology of nucleic acid and nucleic acid fragments, which is applied in the field of recombinant polypeptide production and can solve the problems of time-consuming and expensive
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0439] Cloning of genomic fragments and construction of eukaryotic sIgG / mIgG expression plasmid
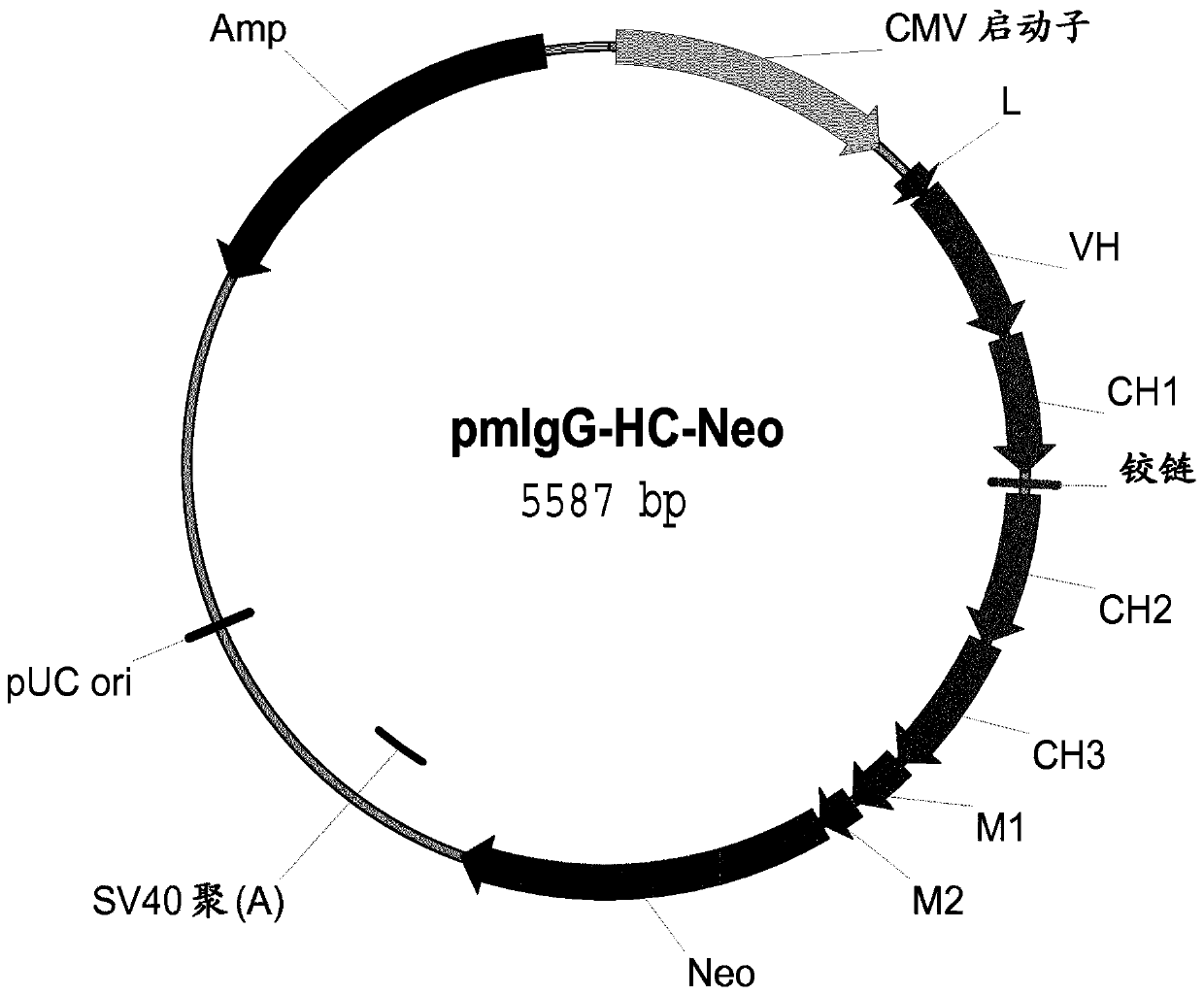
[0440] a) Construction of sIgG expression plasmid pIgG
[0441] In order to express immunoglobulins with binding specificity to protein antigens, plasmids were constructed. It encodes immunoglobulin in the form of secreted sIgG and contains the following elements:
[0442] 1. The transcription unit of human γ1 heavy chain, which contains:
[0443] -Immediate early enhancer and promoter from human cytomegalovirus (hCMV);
[0444] -A synthetic 5'untranslated region containing the Kozak consensus sequence (Kozak, M., Nucleic Acids Res. 15 (1987) 8125-48);
[0445] -Mouse immunoglobulin heavy chain signal sequence, including signal sequence introns;
[0446] -The variable heavy chain cDNA of an antibody with binding specificity to the protein antigen;
[0447] -Including mouse Igμ enhancer (J H 3-J H 4 Transition region) mouse / human heavy chain heterozygous intron 2 which is connected to the huma...
Embodiment 2
[0636] Transfection and library selection
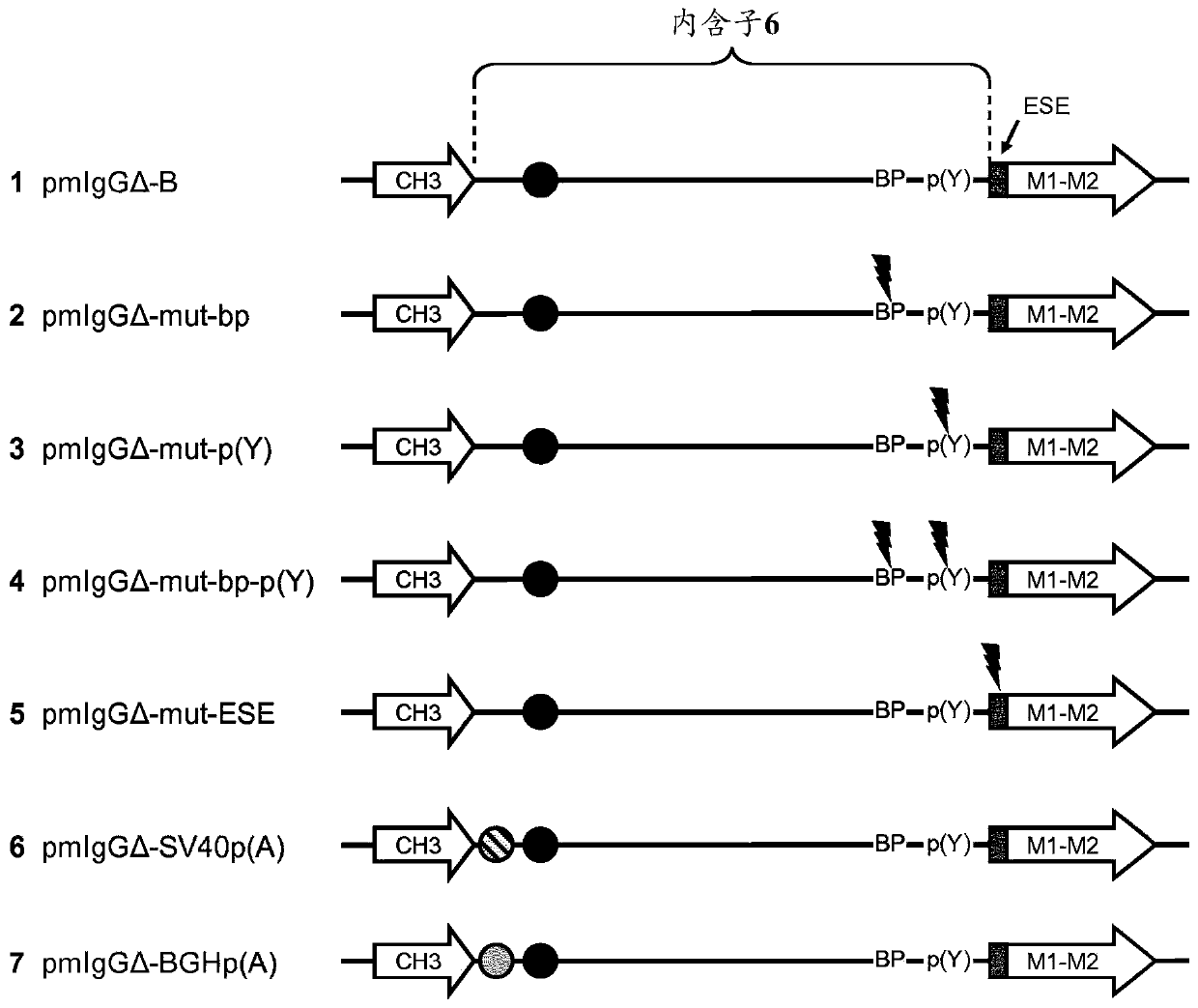
[0637] The CHO-K1 cells (ATCC No. CCL-61; Puck, TT et al., J. Exp. Med. 108 (1958), 945-956) that have been pre-adapted to grow in suspension culture without serum in the suspension culture flask 37°C, 5% CO 2 It was cultured in ProCHO4-CDM medium (Cambrex) supplemented with 8mM L-alanyl-L-glutamine (Invitrogen) and 1x HT supplement (Invitrogen) with 95% humidity. Press the cells 2x10 every 3-4 days 5 Cells / ml are divided into fresh medium. For transfection, press 20μg plasmid DNA / 7.5x10 in a total volume of 200μl PBS at room temperature. 6 Cell electroporation cells. A single 160V pulse, 15ms using a square wave process, with a Gene Pulser XCell electroporator (BioRad) in a 2mm gap electroporation cup for electroporation. The linearized plasmids pmIgGΔ-mut-bp, pmIgGΔ-mut-p(Y), pmIgGΔ-mut-bp-p(Y), pmIgGΔ-mut-ESE, pmIgGΔ-SV40p(A), pmIgGΔ-BGH-p( A) or pmIgGΔ-B transfected cells. Then, one day after transfection, each transfected cell ...
Embodiment 3
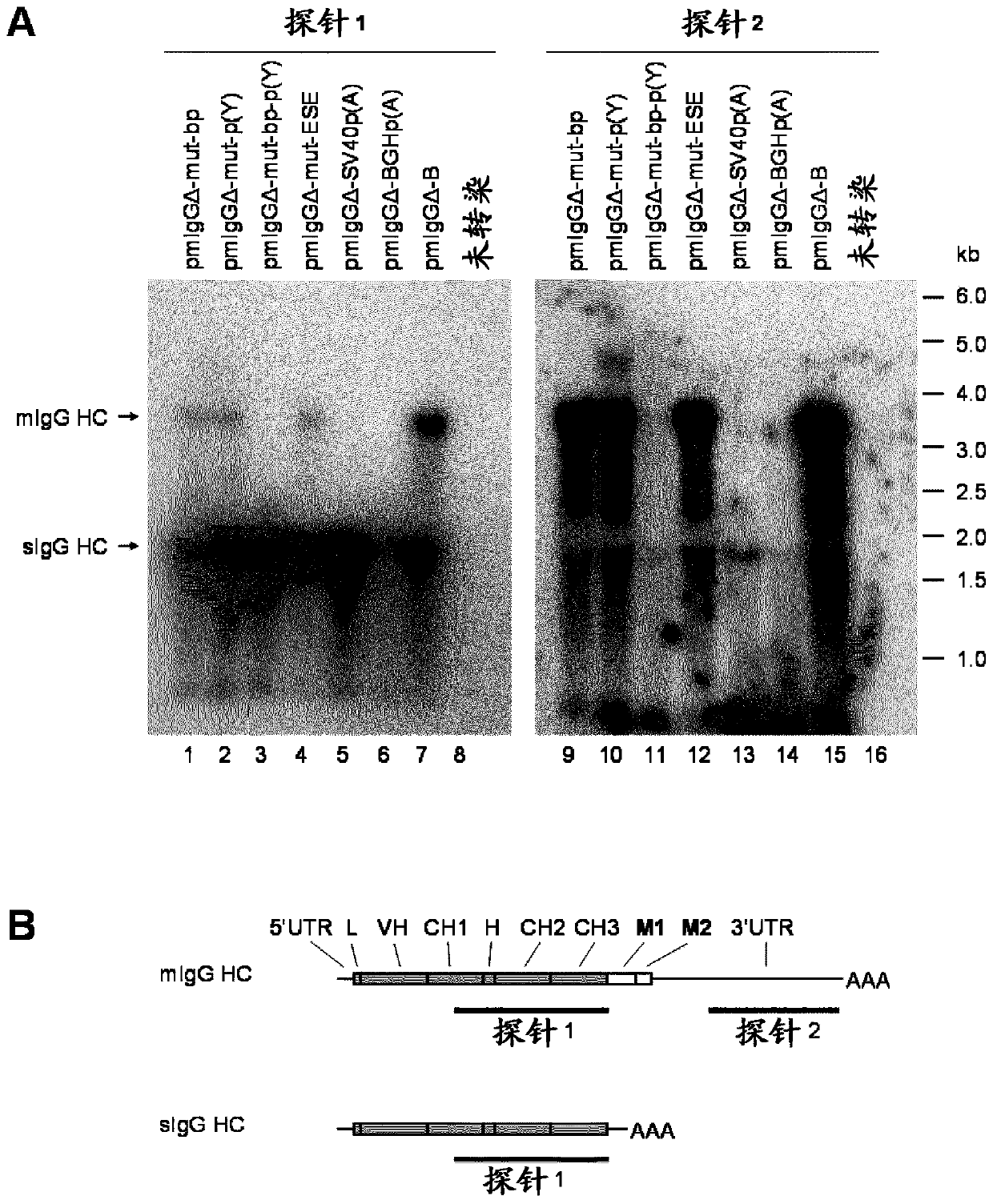
[0639] Northern blot analysis
[0640] In order to determine the ratio between the two heavy chain isoforms arising from the selective RNA processing of the primary transcript, the RNA of the stably transfected clone was analyzed by Northern blot. Therefore, the RNeasy kit (QIAGEN) was used to isolate total RNA from the 7 stably transfected cell banks described in Example 2. In addition, RNA of untransfected CHO-K1 cells was prepared under the same conditions. In each case, 10 μg of total RNA was then fractionated by denaturing agarose gel electrophoresis and transferred to a nylon membrane using the NorthernMax System (Ambion). In order to hybridize the blot membrane, use DECAprime II kit (Ambion) by random priming method with [α- 32 P] labeled two different DNA probes. Such as figure 2 As shown in B, probe 1 and exons CH1 and C H The heavy chain mRNA between 3 is complementary and therefore hybridizes with the two isotypes. In contrast, Probe 2 only binds to the 3'UTR of th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com