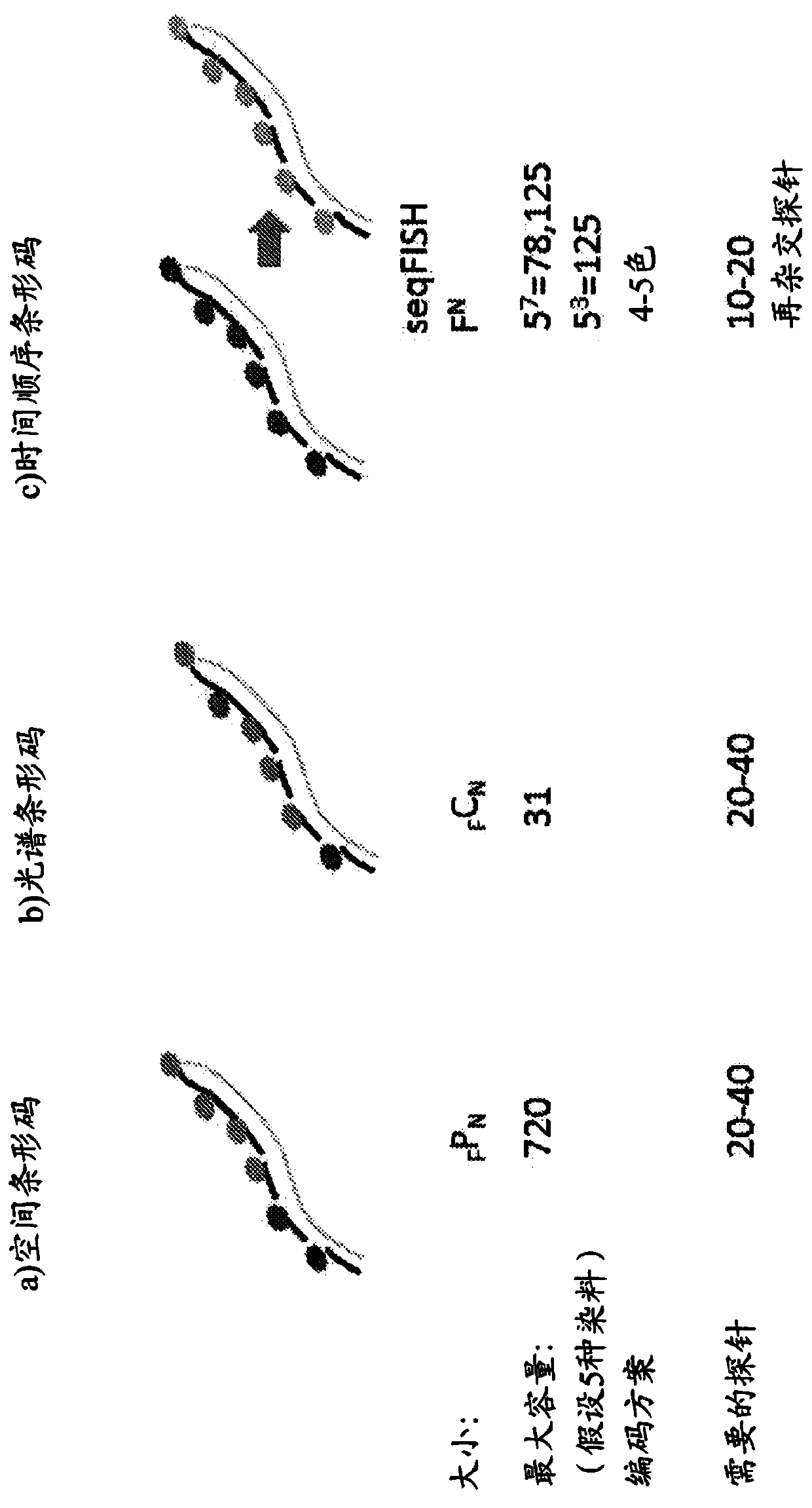
Sequential probing of molecular targets based on pseudo-color barcodes with embedded error correction mechanism
A molecular target and barcode technology, which can be used in the determination/inspection of microorganisms, material excitation analysis, fluorescence/phosphorescence, etc., and can solve problems such as limitations, errors, and introduction bias.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0403] Add barcode based on pseudo-color
[0404] Sample Preparation
[0405] Figure 5 Illustrated are mRNA transcripts immobilized on surfaces by poly-A tails or hydrogel embedding. Once cells are lysed, cell lysates or purified total RNA can be immobilized on the surface by coverslips functionalized with DNA or LNAPoly-T by capturing the poly-A tail of the mRNA. Alternatively, mRNA can be mixed with a hydrogel, such as a polyacrylamide gel, and allowed to gel on the surface of a coverslip, where the pores formed will trap mRNA molecules on the surface.
[0406] Primary Probe Design
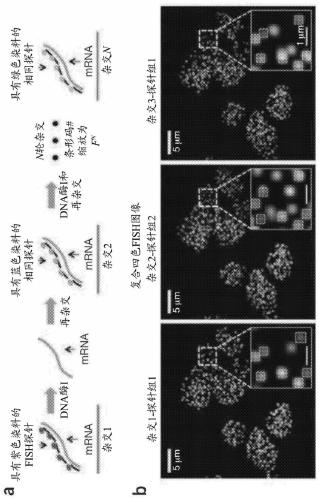
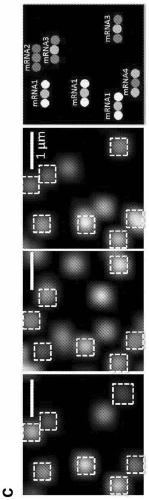
[0407] Figure 6 An exemplary embodiment of gene-specific primary probe design and sequential barcoding hybridization on mRNA immobilized on a surface is illustrated. in particular, Figure 6 a shows that the primary probe is not directly labeled with a fluorophore. The primary probe comprises (i) a gene-specific targeting region of mRNA, which is 20nt to 35nt, (ii) one or more (mult...
Embodiment 2
[0440] Readout probes and rehybridization in mouse embryonic stem cells
[0441] Synthesis of DNA probe-disulfide-dye conjugates
[0442] Exemplary scheme for the synthesis of disulfide-linked readout probe-dye conjugates. Thiol-modified DNA probes in oxidized form were ordered from Integrated DNA Technologies. Ten nanomolar thiol-modified DNA probes were treated with 10 mM TCEP for 30 minutes at 37°C. After the reduction step and gel column purification, the DNA probe was linked with 50 equivalents of N-hydroxysuccinimide 3-(2-pyridyldithio)propionate (SPDP) in a 1x PBS solution containing 10 mM EDTA. mix in. The mixture was allowed to react at room temperature for at least 2 hours. Immediately after the reaction, the mixture was spin column purified and resuspended in 60uL of 1x PBS containing 100ug of cadaverine dye. The reaction was allowed to proceed at room temperature for at least 4 hours before purification by ethanol precipitation and HPLC. The concentration o...
Embodiment 3
[0449] Brain slice analysis
[0450] As an illustration, barcodes generated using the error correction mechanism disclosed herein were used for in situ transcriptional analysis of single cells, revealing the spatial organization of cells in the mouse hippocampus.
[0451] Identifying the spatial organization of tissues at cellular-level resolution from single-cell gene expression profiles is critical for understanding many biological systems. In particular, there is conflicting evidence as to whether the hippocampus is organized into transcriptionally distinct subregions. Here, a generalizable in situ 3D multiplex imaging approach was applied to quantify hundreds of genes at single-cell resolution by sequentially barcoded fluorescence in situ hybridization (seqFISH) (Lubeck et al., 2014). seqFISH was used to identify unique transcriptional states by quantifying and clustering up to 249 genes in 16,958 cells. By visualizing these clustered cells in situ, we identified distinc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com