Novel circRNA and application thereof
A sequence and reagent technology, applied in the field of disease detection, can solve the problems of lack of effective methods for early diagnosis and achieve good application prospects, good stability, fast and accurate quantitative results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The establishment and sampling of the hypoxia-ischemia rat model of embodiment 1
[0035] 7-day-old neonatal SD rats with a body weight of 10-14 g. They were randomly divided into two groups, a sham operation group (normal control group) and a hypoxic-ischemic group, with 3 rats in each group. Under ether anesthesia, a median neck incision was made, and the left common carotid artery was separated. The sham operation group did not receive any treatment; the operation group ligated and cut off the left common carotid artery, rested for 1 hour, and placed in 8% oxygen and nitrogen mixed gas for 2 hours. One day after the experiment, the rats in each group and at each time point were decapitated under ether anesthesia, and the brain tissues were taken out and immediately put into liquid nitrogen for cryopreservation.
Embodiment 2
[0036] Example 2 High-throughput sequencing and data analysis
[0037] 2.1 Extraction and quality inspection of tissue total RNA
[0038] (1) Cut the tissues of each group into pieces on ice, take an appropriate amount (50-100 mg) of the tissues, add 1 ml of pre-cooled Trizol to the EP tube without RNase, pipette evenly, and let stand on ice for 10 minutes;
[0039] (2) Add 200 μl of pre-cooled chloroform, shake vigorously for 30 sec, then let stand on ice for 15 min, and centrifuge at 14000 rpm / min at 4°C for 20 min;
[0040] (3) Take out the supernatant and put it in a new RNase-free EP tube (be careful not to touch the middle layer), add an equal volume of pre-cooled isopropanol (about 500 μl), mix well and let stand on ice for 0.5-1h, 4 Centrifuge at 14000rpm / min for 20min;
[0041] (4) Discard the supernatant, add 1 ml of pre-cooled 75% ethanol, mix upside down, and centrifuge at 14000 rpm / min at 4°C for 20 min;
[0042] (5) Discard the supernatant, add 1 ml of pre-coo...
Embodiment 3
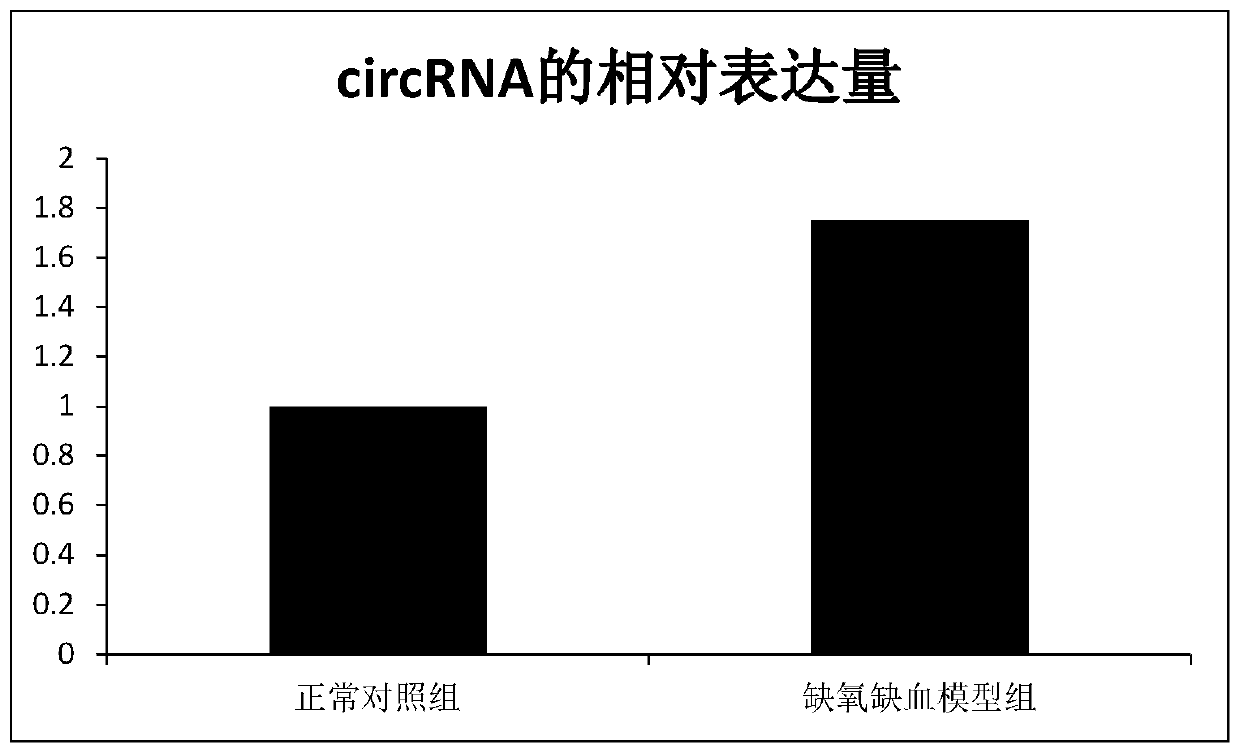
[0062] Example 3 RT-PCR verification of the relative expression of circRNA in the control group and the model group
[0063] 1. Model building
[0064] Concrete method is with embodiment 2. The sham operation group (normal control group) and the hypoxia-ischemia group had 20 rats in each group.
[0065] 2. Experimental method
[0066] 2.1 Extraction of total tissue RNA
[0067] Reference Example 2
[0068] 2.2 Primer design
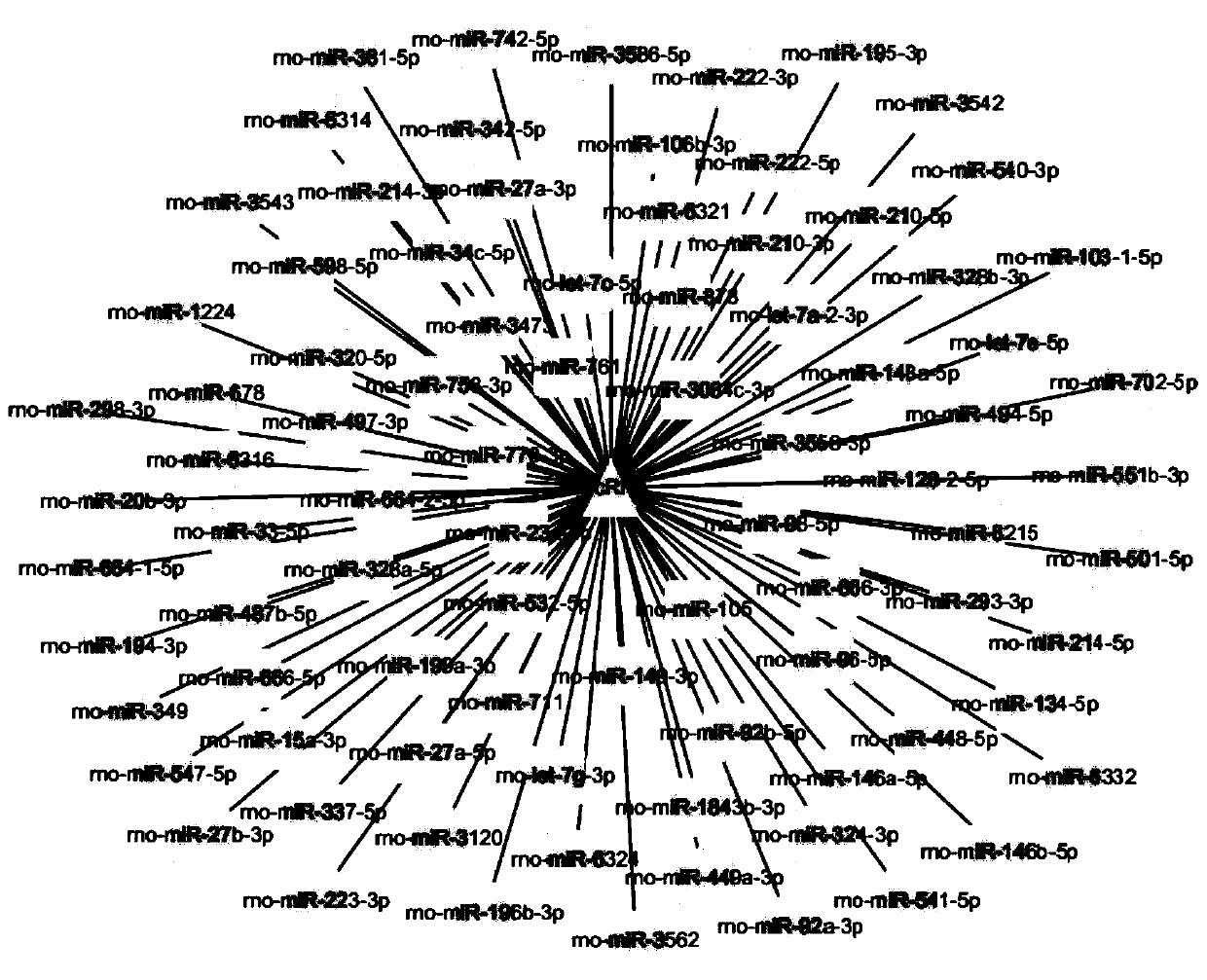
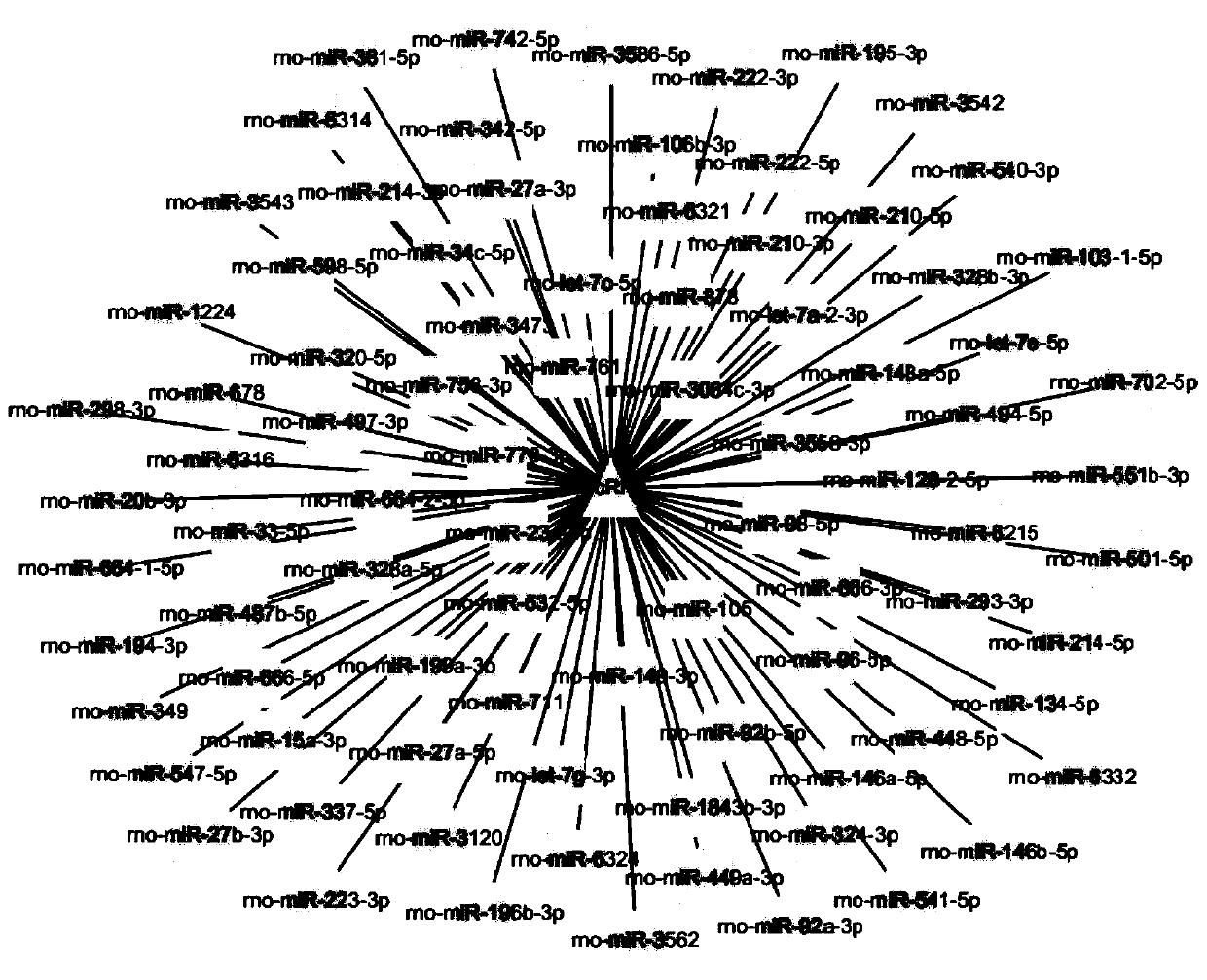
[0069] For the circular closed structure of circRNA, design back-to-back primers (divergent primer), GAPDH is used as an internal reference, and sent to a primer synthesis company for synthesis.
[0070] circRNA primers:
[0071] Upstream primer: 5'-CAGTCAGATGGTGGTTCATTTCA-3' (SEQ ID NO.2)
[0072] Downstream primer: 5'-GGTCATAGAAACCCAAAAGTGC-3'(SEQ ID NO.3)
[0073] Target gene amplification length: 145bp.
[0074] 2.3 Reverse transcription
[0075] According to PimeScript TM RT reagent Kit (Perfect Real Time) RR037A (Takara) manual configure...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com