Preparation method and application of humanized CD3 genetically-modified animal model
A humanized, non-human animal technology, applied in the field of biomedicine, can solve the problems of cumbersome model building process, low accuracy, high cost, reduce drug development risks, speed up the research and development process, save time and cost. cost effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0166] Embodiment 1 sequence design (partial replacement)
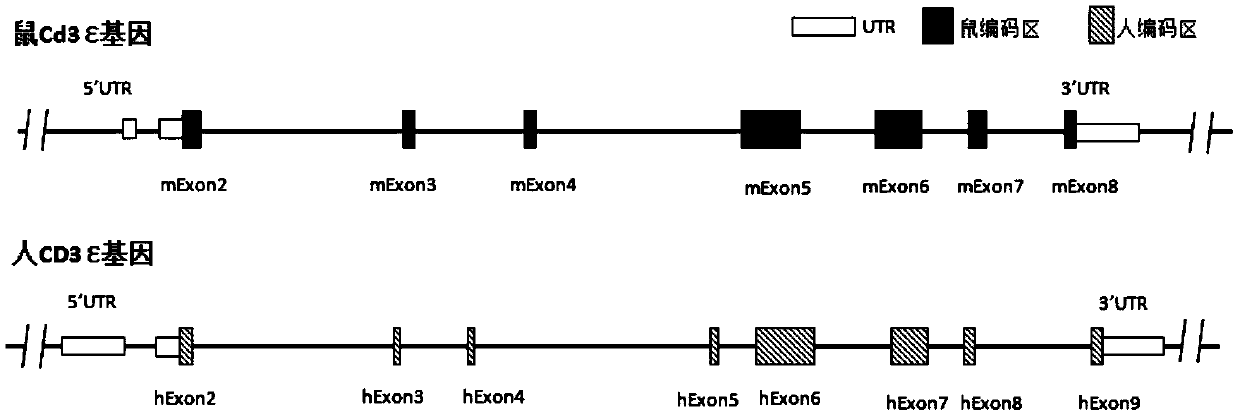
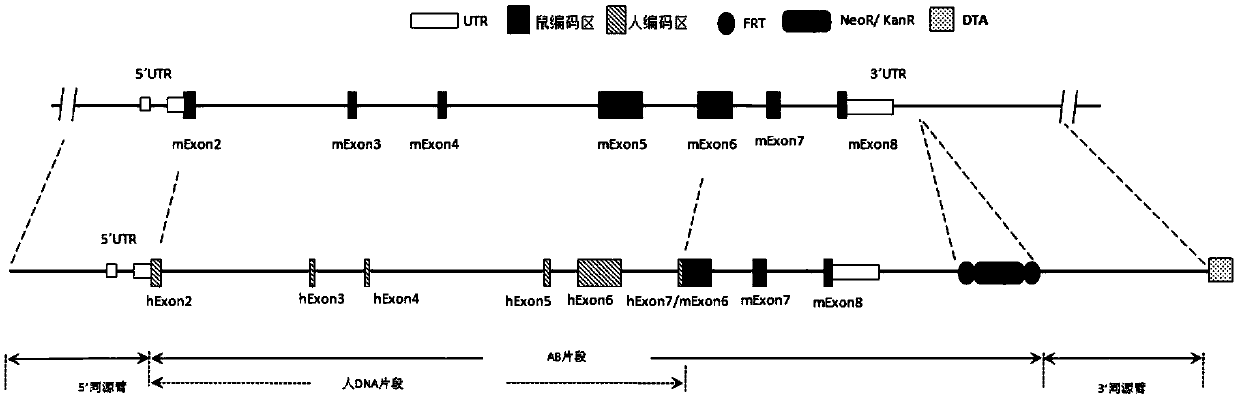
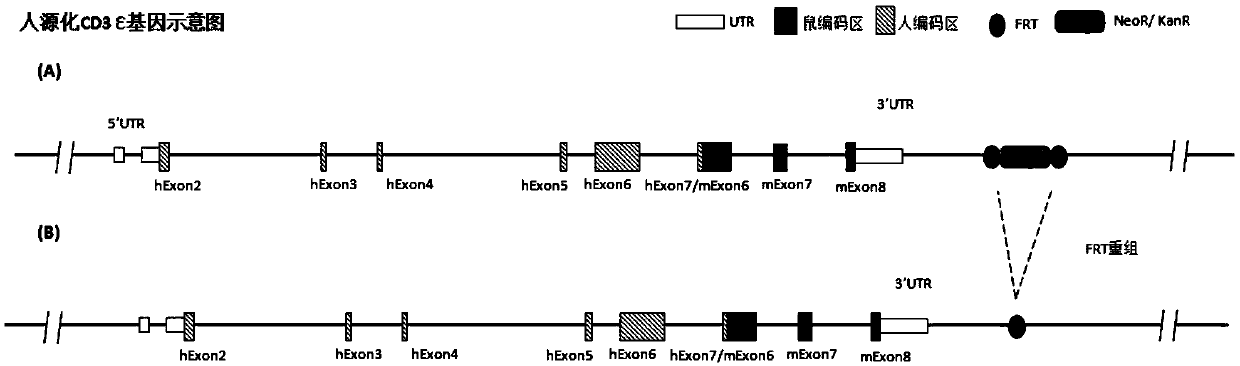
[0167] According to the purpose of the invention, designed figure 2 The targeting strategy for partial sequence replacement of the Cd3ε gene and the design of the targeting vector are shown. The targeting vector includes 5' homology arm (SEQ ID NO: 1), 3' homology arm (SEQ ID NO: 2) and human DNA fragment (SEQ ID NO: 3), and was constructed on the recombinant vector for positive cloning The screened resistance gene, such as the neomycin phosphotransferase coding sequence Neo, is equipped with two site-specific recombination systems arranged in the same direction on both sides of the resistance gene, such as the Frt recombination site (or conventional LoxP recombination system). Further, a gene encoding a negative selection marker, such as the gene encoding diphtheria toxin A subunit (DTA), was constructed downstream of the 3' homology arm of the recombinant vector. image 3 A schematic diagram of the humanized mou...
Embodiment 2
[0168] Example 2 Homologous Recombination Fragment Primer Design and PCR Amplification
[0169] Primers were designed to amplify 7 homologous recombination fragments (A1, A2-1, A2-2, A3, B, C1, C2). The primer sequences are shown in Table 1.
[0170] Table 1 Homologous recombination fragments and their corresponding lengths and primer sequences
[0171]
[0172] Using BAC as a template, 7 homologous recombination fragments were amplified using KOD-plus-polymerase. Among them, A1, A3, B, C1, and C2 selected the bacterial artificial chromosome (Bacterial Artificial Chromosome, BAC) (number: RP23-140I1, hereinafter referred to as "bacterium containing mouse BAC") containing the mouse Cd3ε genome as a template, and A2- 1. A2-2 selects the bacterial artificial chromosome containing the human CD3ε genome (number: RP11-414G21, hereinafter referred to as "bacteria containing human BAC") as a template. The specific PCR reaction system and reaction conditions are as follows:
[01...
Embodiment 3
[0178] Example 3 Construction of Homologous Recombination Targeting Vector
[0179] The targeting vector construction process is as follows:
[0180] 1. Ligate the fragment B obtained in Example 2 to the pBs-Neo vector by enzyme-cut ligation (BamHI / NotI) to obtain the pBs-Neo-B plasmid, and send it to the sequencing company for verification;
[0181] 2. After connecting the fragments A1 and A2-1, A2-2 and A3 obtained in Example 2 using overlap PCR (phusion enzyme) (see Table 4 and 5 for the reaction system and conditions), send them to the sequencing company for sequencing to confirm A1+A2 After the -1 and A2-2+A3 fragments are connected correctly, the fragments are digested and connected to the pBs-Neo-B plasmid (HindIII / SwaI / XhoI) to obtain pBs-Neo-(A1+A2-1+A2-2+ A3+B) plasmid;
[0182] 3. The pBs-Neo-(A1+A2-1+A2-2+A3+B) plasmid was electrotransformed into bacteria containing human BAC to obtain the pBs-Neo-(AB) plasmid containing the AB fragment; the AB fragment (see fi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com