Identification method of sex-linkage Luhua chicken genotype of Chinese native chickens
A chicken breed and Chinese technology, which is applied in the identification field of the sex-linked Luhua chicken genotype of native chicken breeds in my country, can solve the problems of unclear function of CDKN2A gene, undetermined specific mutation sites of phenotype, and deletion, etc., and achieves good application. Value, high accuracy, simple operation effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
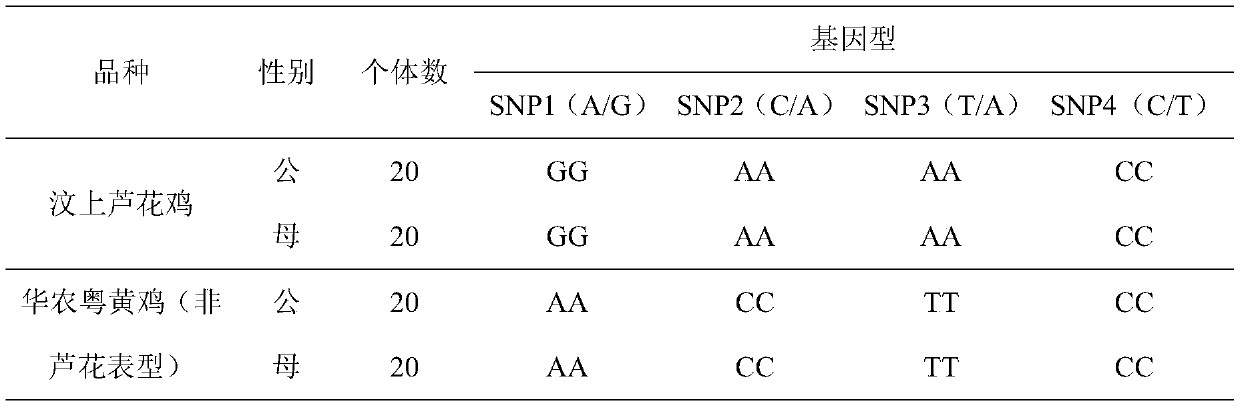
[0037] Embodiment 1 A kind of method of detecting chicken sex-linked reed flower phenotype
[0038] SNP1 is the CDKN2A gene promoter region and 267bp upstream of the transcription initiation site. Primers were designed according to its position on the genome, and primers with high amplification specificity were screened out, and the amplification system and reaction program were optimized.
[0039] 1. Experimental method
[0040] Collect chicken blood samples, extract blood sample DNA, and amplify the blood sample DNA. The amplification primers are:
[0041] SNP1 upstream primer: 5'-GTTACCCGTTTCTGCTCCTTGACGC-3' (SEQ ID NO: 1);
[0042] SNP1 downstream primer: 5'-GCCCCCACCGTAAGAAAGCTCTCTC-3' (SEQ ID NO: 2).
[0043] The 50μL amplification system is as follows:
[0044]
[0045] SNP1 amplification program: pre-denaturation at 94°C for 2 minutes; denaturation at 98°C for 10 seconds, annealing at 60°C for 20 seconds, extension at 68°C for 50 seconds, a total of 40 cycles; ex...
Embodiment 2
[0049] Embodiment 2 A kind of method of detecting chicken sex-linked reed flower phenotype
[0050] SNP2 is the 380bp of intron 1 of CDKN2A gene. According to its position on the genome, primers were designed, and primers with high amplification specificity were screened out, and the amplification system and reaction program were optimized.
[0051] 1. Experimental method
[0052] Collect chicken blood samples, extract blood sample DNA, and amplify the blood sample DNA. The amplification primers are:
[0053] SNP2 upstream primer: 5'-GGTCGGCAGGCTATCTCTAT-3' (SEQ ID NO: 3);
[0054] SNP2 downstream primer: 5'-AGGTCCACGTATTCTCATCA-3' (SEQ ID NO: 4).
[0055] The 50μL amplification system is as follows:
[0056]
[0057] SNP2 amplification program: pre-denaturation at 94°C for 3 minutes; denaturation at 94°C for 30 seconds, annealing at 58°C for 30 seconds, extension at 72°C for 30 seconds, a total of 34 cycles; extension at 72°C for 5 minutes.
[0058] After purification,...
Embodiment 3
[0061] Embodiment 3 A kind of method of detecting chicken sex-linked reed flower phenotype
[0062] SNP3 is at 170bp of exon 1 of CDKN2A gene, and primers were designed according to its position on the genome, and primers with high amplification specificity were screened out, and the amplification system and reaction program were optimized.
[0063] 1. Experimental method
[0064] Collect chicken blood samples, extract blood sample DNA, and amplify the blood sample DNA. The amplification primers are:
[0065] SNP3 upstream primer: 5'-CTCTCCTGTTCCCATGACCTCTCG-3' (SEQ ID NO: 5);
[0066] SNP3 downstream primer: 5'-GATAGAGATAGCCTGCCGACCACC-3'(SEQ ID NO:6),
[0067] The 40μL amplification system is as follows:
[0068] Template DNA 1.5 μL
[0069] 10pmol primers 1.8μL each
[0070] 1.1×T3 Super PCR Mix 35μL
[0071] SNP3 amplification program: pre-denaturation at 98°C for 3 minutes; denaturation at 98°C for 10 seconds, annealing at 65°C for 10 seconds, extension at 72°C for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com