Simple and convenient method and process for detecting SMA by DNA sequencing
A technology of DNA sequencing and sequencing, which is applied in biochemical equipment and methods, microbial measurement/inspection, etc., and can solve problems such as cumbersome detection process and difficulty in mastering by ordinary medical units
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
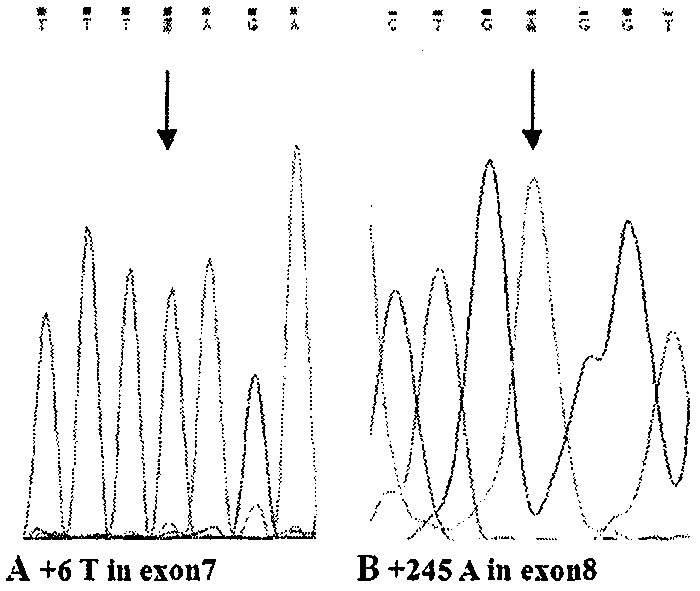
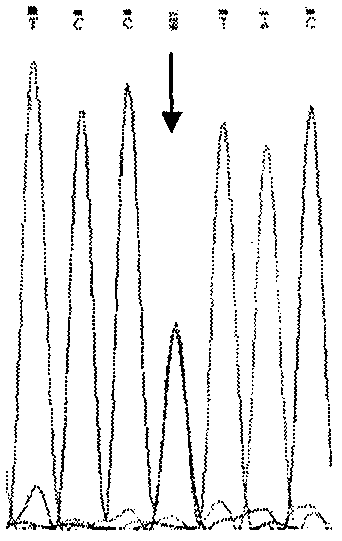
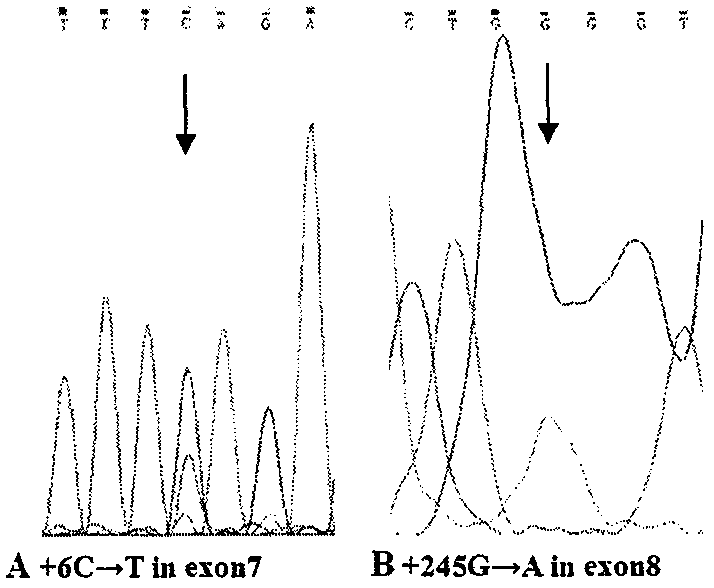
[0011] We performed PCR amplification on SMN gene DNA inzygote 6-exon 8, and sequenced and analyzed the PCR amplification products, summed up and explored a simple method and process for DNA sequencing to detect SMA.
[0012] 1. PCR amplification
[0013] 1. Primer sequence and synthesis
[0014] Intrazygote 6-exon 8 of the SMN gene was amplified by PCR and synthesized by Invitrogen. The primer sequences, amplified fragments and annealing temperatures are shown in Table 1.
[0015] Table 1, primer conditions and products
[0016]
[0017] 2. PCR reaction system
[0018] The total reaction volume is 25 μl, including:
[0019]
[0020] Addition of autoclaved ddH 2 0 to 25.0 μl
[0021] The preparation of the PCR reaction system was performed on ice, and finally Taq DNA polymerase was added, mixed evenly and distributed.
[0022] 3. PCR reaction conditions
[0023] The PCR amplification conditions were pre-denaturation at 94°C for 7 minutes, 94°C for 45 sec, 56°C for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com