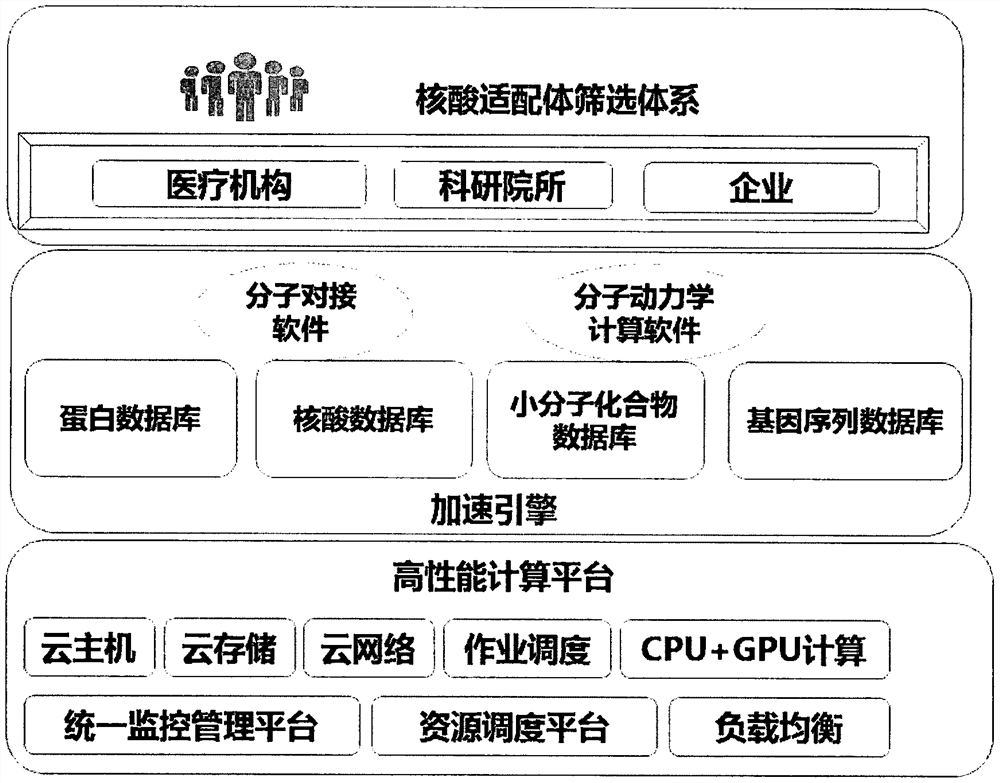
A high-performance computing platform-based computer-aided screening method for nucleic acid aptamers and nucleic acid aptamers
A nucleic acid aptamer and computer-aided technology, which is applied in the field of molecular biology detection, can solve the problems of complex screening process, time-consuming, and many times of realization, and achieve the effect of improving screening efficiency, reducing the number of experiments, and reducing the number of experiments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0118]Example 1: When the conformation of the protein A binding nucleic acid A, the energy is -6 kcal / mol when the B conformation is combined, then the binding the B conformation is stable, and this value is generally -3kcal. / mol or less is preferred, therefore, in general, the B conformation is desirable.
Embodiment 2
[0119]Example 2: When the conformation of the protein A binding nucleic acid A is -5 kcal / mol, the energy is -6 kcal / mol when the B conformation is binding, but the main hydrophobic portion in the B conformation is bonded to the hydrophilic end of the protein A, then Even if the energy of the B conformation is low, the B conformation is not advisable.
Embodiment 3
[0120]Example 3: The energy is -5 kcal / mol, and the energy is -6 kcal / mol, but the main hydrophilic portion in the B conformation is combined in the hydrophilic end of the protein A in the primary hydrophilic portion of the B conformation. Then the energy of the B conformation is low, and the main hydrophilic portion in the B conformation is bonded to the hydrophilic end of the protein A, and therefore the B conformation is preferable.
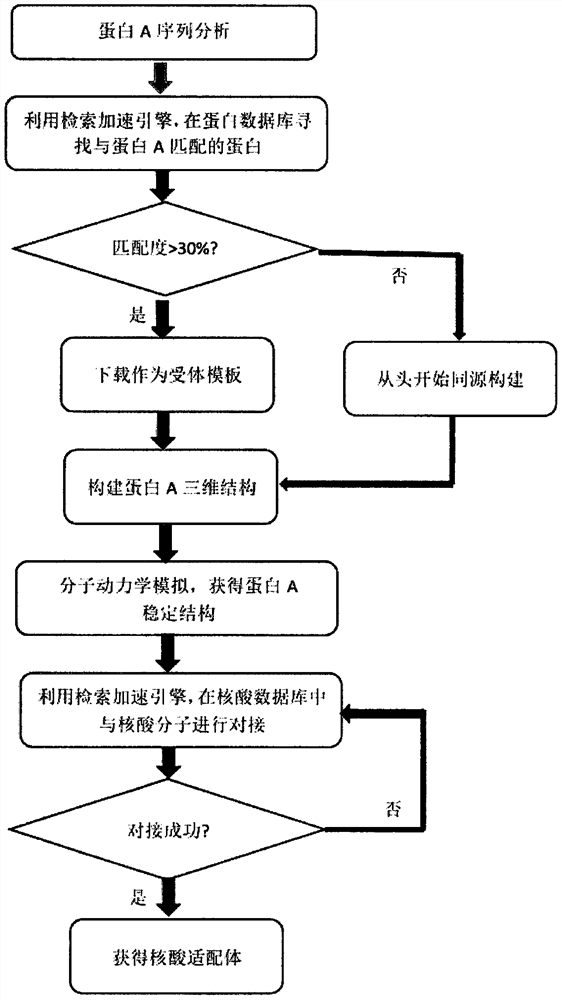
[0121]Always, in the specific embodiment of the present invention, a nucleic acid adaptation computer assist screening method based on a high performance computing platform is capable of querying whether to match the protein structure of the target protein according to the matching principle, if it is Body template, if the homologous construction is not performed, the nucleic acid adapter is obtained by molecular dynamics analog and molecular docking screening, the computer auxiliary screening method is less experiment, efficient and fast, can be a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com