LAMP primer combination for detecting 5 gram-positive bacteria in intraocular fluid and application thereof
A primer combination and primer set technology, applied in the biological field, can solve the problems of easy contamination, long PCR detection time and high false positive rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0156] Embodiment 1, the preparation of kit
[0157] The kit consists of five LAMP primer sets, each for the detection of 1 Gram-positive bacterium in intraocular fluid.
[0158] The primer set used to detect Staphylococcus epidermidis is as follows (5'→3'):
[0159] Outer primer F3 (SEQ ID NO: 1): ATTgAgATAgCgggggA;
[0160] Outer primer B3 (SEQ ID NO: 2): ACAACAAAgTAACAgTACCATg;
[0161] Internal primer FIP (SEQ ID NO: 3): gCgTCATgCCTTTTATTTgAAgAAAATgTACAgTCATAgCTAgTggA;
[0162] Internal primer BIP (SEQ ID NO: 4): ACAggAgTAAATTCAgTgATTgCAATTCCgCAACTTACAAAACATg;
[0163] Loop primer LF (SEQ ID NO: 5): TTATATgTATgTgCCCAAAATCACA;
[0164] Loop primer LB (SEQ ID NO: 6): CCAATTgATTggAAAggATTTgATC.
[0165] The primer set used to detect Staphylococcus aureus is as follows (5'→3'):
[0166] Outer primer F3 (SEQ ID NO: 7): gCAACTgAAACAACAgAAgC;
[0167] Outer primer B3 (SEQ ID NO: 8): TTTTgTgTTgggCgAgC;
[0168] Internal primer FIP (SEQ ID NO: 9): TCACggATACCTgTACCAgCATCCTC...
Embodiment 2
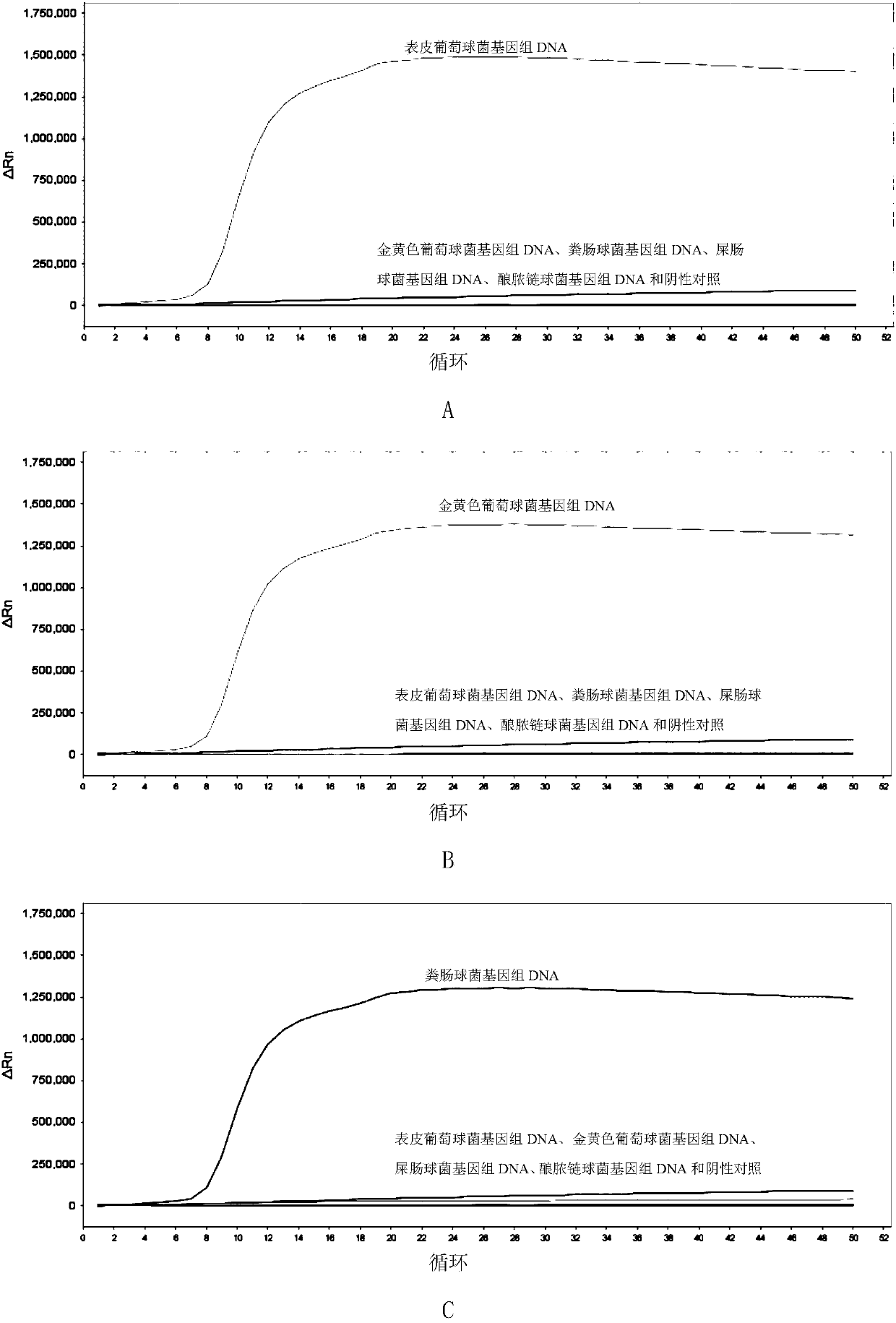
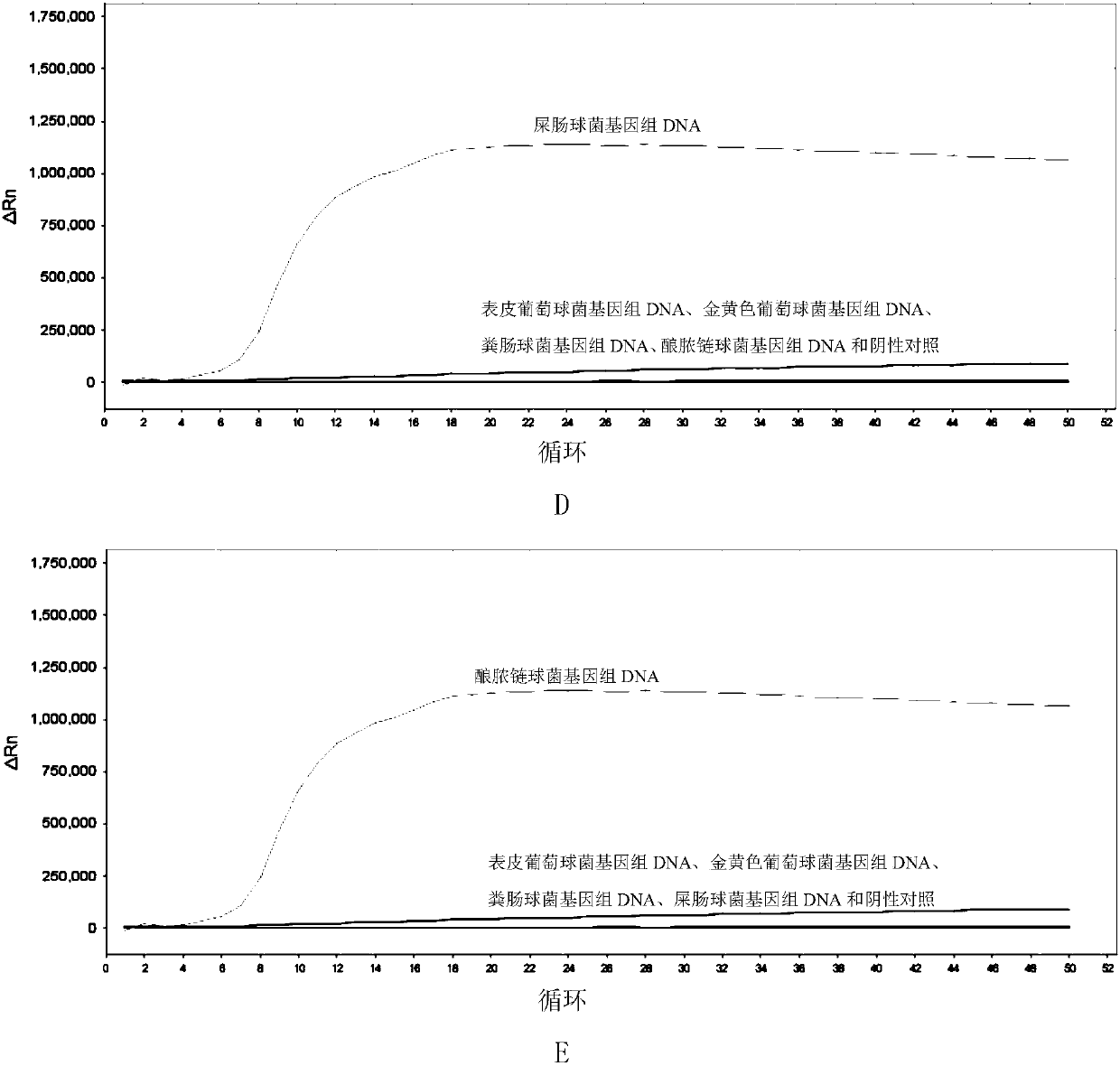
[0193] Embodiment 2, specificity
[0194] Test sample 1: Staphylococcus epidermidis.
[0195] Test sample 2: Staphylococcus aureus.
[0196] Test sample 3: Enterococcus faecalis.
[0197] Test sample 4: Enterococcus faecium.
[0198] Test sample 5: Streptococcus pyogenes.
[0199] Each sample to be tested carries out the following steps respectively:
[0200] 1. Extract the genomic DNA of the sample to be tested.
[0201] 2. Using the genomic DNA extracted in step 1 as a template, each primer set prepared in Example 1 was used to perform loop-mediated isothermal amplification.
[0202] Reaction system (10 μL): 1 μL 10×ThermoPol Buffer, 1.6 μL betaine with a concentration of 5M, 0.1 μL BSA aqueous solution with a concentration of 50 mg / mL, and 0.4 μL MgSO with a concentration of 100 mM 4 Aqueous solution, 0.3 μL 20×EvaGreen, 0.15 μL of 100 mM dNTPs (each), 0.4 μL of 8 U / mL Bst DNA polymerase large fragment, genomic DNA of the sample to be tested (between 50 pg and 50 ng),...
Embodiment 3
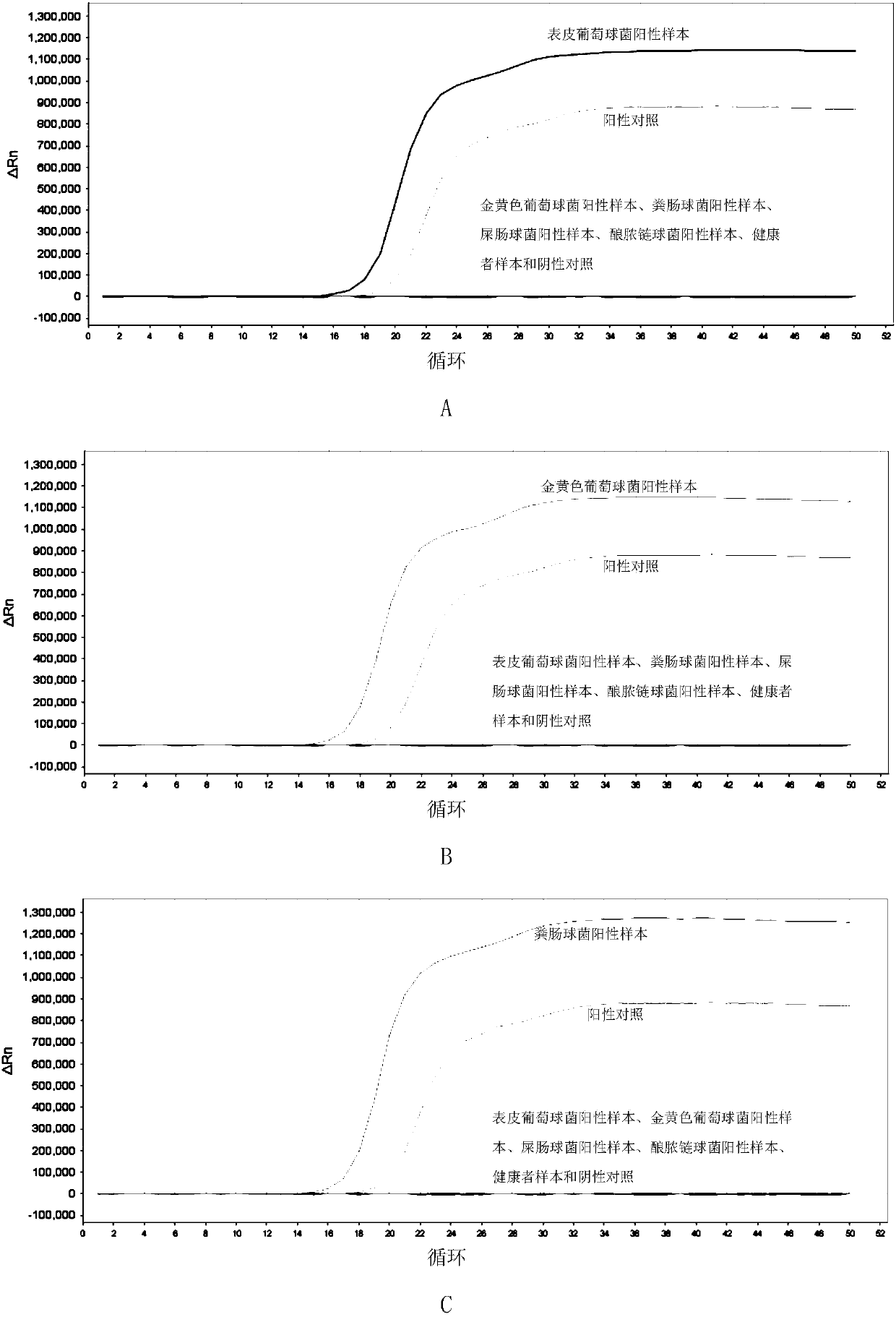
[0212] Embodiment 3, sensitivity
[0213] Test sample 1: Staphylococcus epidermidis.
[0214] Test sample 2: Staphylococcus aureus.
[0215] Test sample 3: Enterococcus faecalis.
[0216] Test sample 4: Enterococcus faecium.
[0217] Test sample 5: Streptococcus pyogenes.
[0218] Each sample to be tested carries out the following steps respectively:
[0219] 1. Extract the genomic DNA of the sample to be tested, and perform gradient dilution with sterile water to obtain each dilution.
[0220] 2. Using the dilution obtained in step 1 as a template, the primer sets prepared in Example 1 were used to perform loop-mediated isothermal amplification.
[0221] When the sample to be tested is the sample to be tested 1, the loop-mediated isothermal amplification is performed using primer set I. When the sample to be tested is the sample to be tested 2, loop-mediated isothermal amplification is performed using primer set II. When the sample to be tested is the sample to be test...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com