Biological information quality control method and device based on next-generation sequencing and storage medium
A technology of biological information and quality control methods, which is applied in the fields of biological information quality control methods, devices and storage media based on next-generation sequencing, which can solve the problems of great influence on the accuracy of test results, low detection sensitivity of low-frequency mutations, and large sample consumption and other issues to achieve the effect of avoiding errors in subsequent mutation detection results, avoiding false positive results, and avoiding cost problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
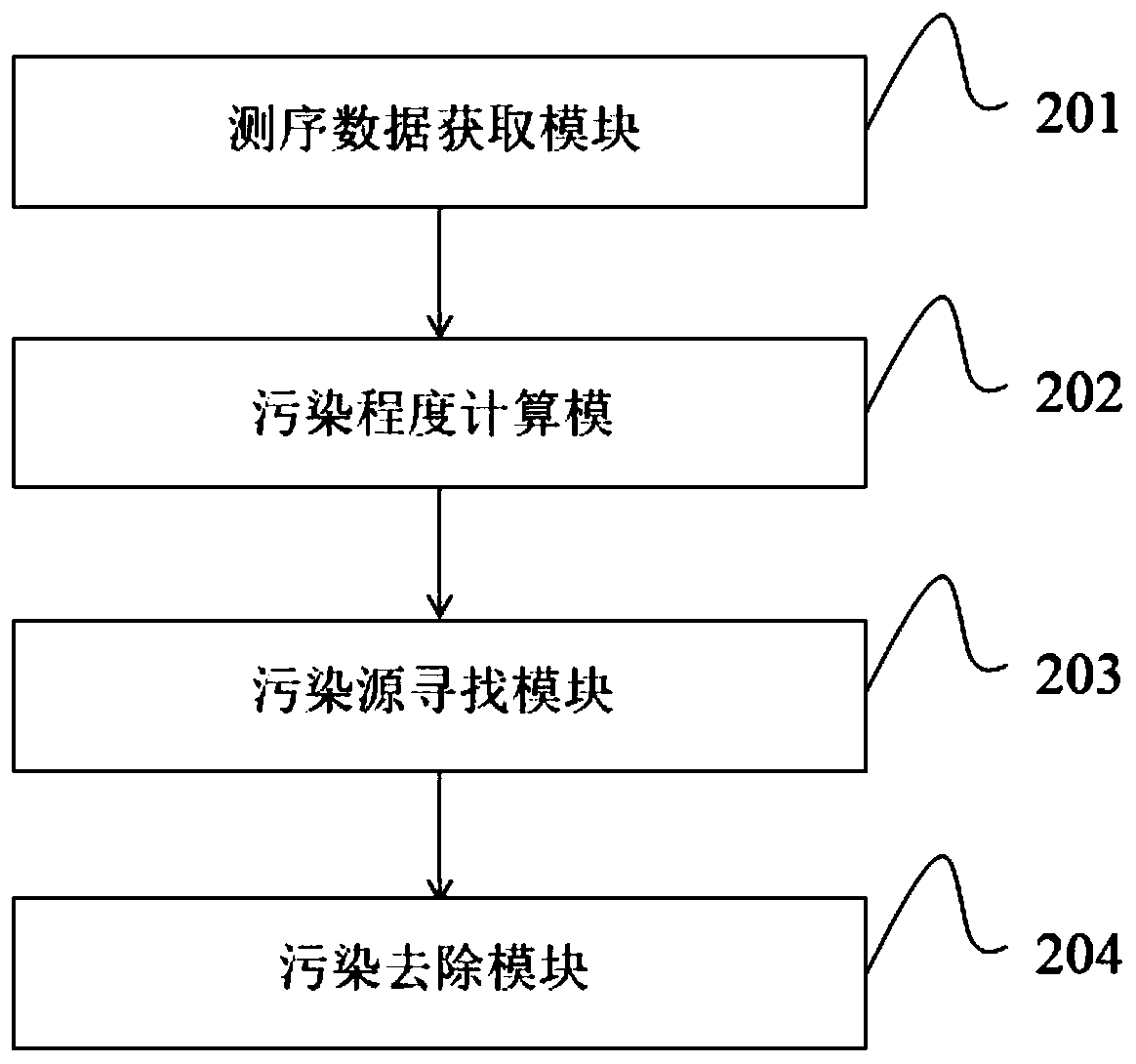
Image
Examples
Embodiment 1
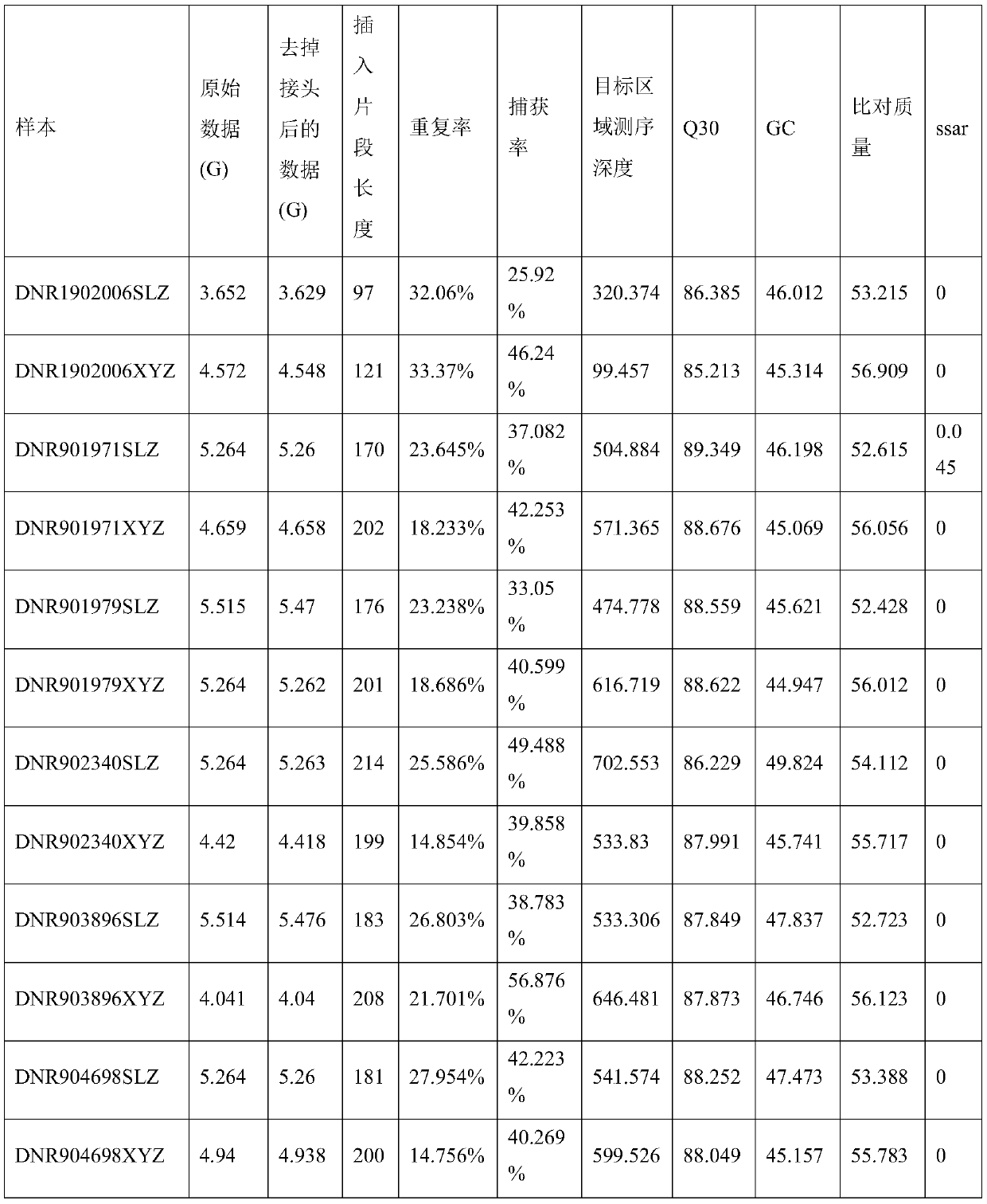
[0063] In this example, 6 pairs of paired samples (white blood cells + tissue samples) were compared for batch sample quality control information. The comparison results are shown in Table 1. It can be known that the capture efficiency and insert length of the sample DNR1902006 SLZ are significantly lower than those of the same batch. For the samples of the experimental method, the sample DNR1902006SLZ can be judged as a quality control unqualified sample through the batch sample quality control information comparison method of the present invention, and the sample degradation can be further determined. In the subsequent copy number variation detection, in the sample coverage normalization step, due to the low capture efficiency, the coverage of the target region after normalization was low, resulting in the missing detection of many copy number variations. From the quality control information, it can be judged that these copy number variations cannot give results and are false...
Embodiment 2
[0067] In this embodiment, the samples used are CT1900260XYZAA03 (sample number) and the corresponding white blood cell control sample DN1900260XYZAA03 (sample number). In this example, a problem was found in the contamination quality control of the sample. There are 16 homozygous quality control sites in the sequencing data of the control samples, and 8 of these homozygous sites are non-homozygous sites in the tissue samples, which are regarded as contamination sites. Calculate the average of the mutation frequencies of these 8 non-homozygous sites, and get the pollution degree of the sample in this case to be 24%, which is greater than the pollution threshold of 1%. It is determined that the sample in this case is contaminated, and then find the samples in the same batch including these 8 The contamination source DN1900852SLZAA01 (sample number) of non-homozygous sites, and after removing all the mutations of the contamination source, the correct mutation detection result of...
Embodiment 3
[0069] In this embodiment, the samples used are CT1901812XYZAA01 (sample number) and the corresponding white blood cell control sample DN1901812XYZAA01 (sample number). In this example, a problem was found in the contamination quality control of the sample. There are 18 homozygous quality control sites in the sequencing data of the control sample, and these homozygous sites have 6 non-homozygous sites in the tissue samples, which are regarded as pollution sites, and these 6 non-homozygous sites are calculated The average value of site mutation frequency shows that the pollution degree of the sample in this case is 5%, which is greater than the pollution threshold of 1%. It is determined that there is contamination in the sample of this case, and then no samples including these 6 non-homozygous sites can be found in the same batch of samples For the sample points, a total of 192 mutations with a mutation frequency of less than 5% in the sample and belonging to the known populat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com