Glioma EM/PM molecular typing method based on Taqman low-density chip and application of glioma EM/PM molecular typing method
A glioma and genetic technology, applied in biochemical equipment and methods, microbial measurement/testing, instruments, etc., can solve the problems of stagnant research on molecular typing and has not yet been transformed into individualized diagnostic tools
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0082] Example 1. Establishment of EM / PM molecular typing of training sample library
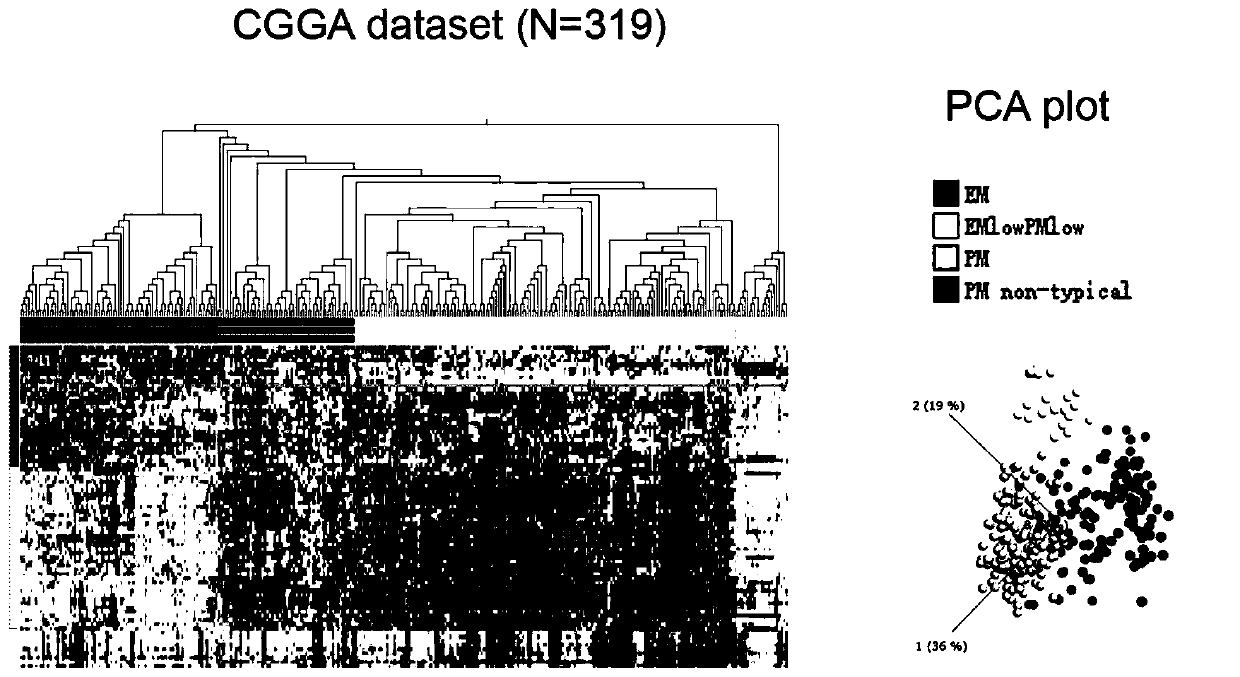
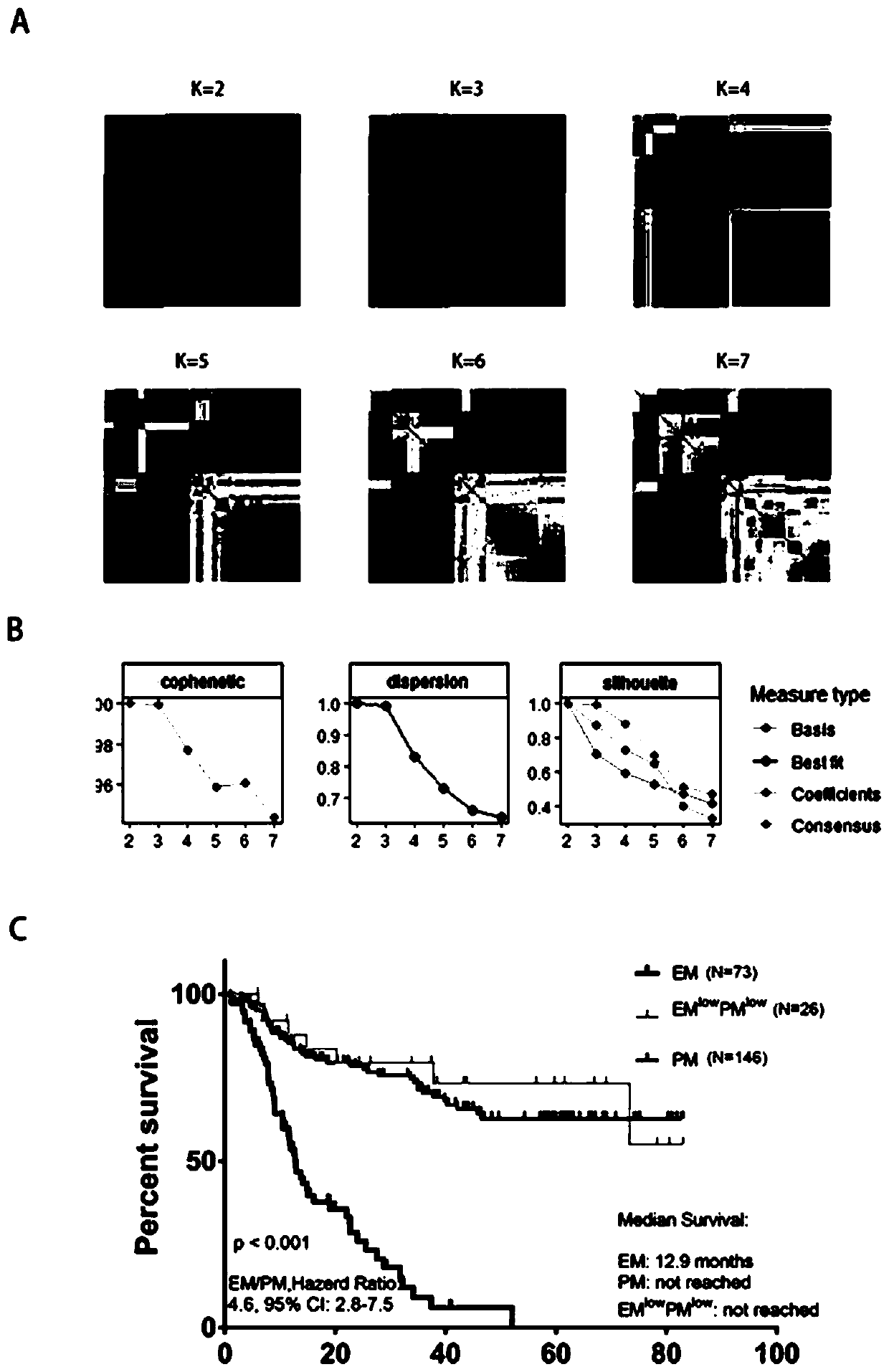
[0083] The goal of the present invention is to achieve EM / PM molecular typing of individual patients. To achieve this goal, a supervised classification algorithm is required, and a supervised classification algorithm requires a set of known samples as a training sample set. . The Chinese Glioma Genome Atlas (CGGA) database contains the mRNA sequencing data of 319 glioma samples. Some of the above samples still have tumor tissues, which can be used for RNA extraction and TLDA chip detection. The data obtained is used as Establish training set of individualized EM / PM typing prediction model.
[0084] First, through the analysis of the public brain glioma gene expression database REMBRADNT, the coefficient of variation (CV) of candidate EM / PM classification genes was calculated, and a total of 27 EM genes, 32 PM genes, and 9 markers with large coefficient of variation were selected. EM low PM low ...
Embodiment 2
[0087] Example 2. Screening of glioma typing gene group and preparation of detection chip
[0088] 1. Screening of glioma typing gene group
[0089] Using the genes described in Table 1, a panel containing 96 genes was designed.
[0090] Table 1
[0091]
[0092]
[0093]
[0094]
[0095] These genes were handed over to the company to design primers and prepare Taqman low-density chips. In order to preliminarily evaluate the actual performance of the platform and further narrow the range of candidate genes based on the real-time fluorescent quantitative PCR data, 40 typical glioma samples among the 263 samples described in Example 1 were selected and tested using the chip . It was found that among the 96 genes, 6 genes (DMRTA2, SHOX2, SOX4, TNFRSF19, MOBP, POLR2F) had large CT values or were not amplified in most samples. These genes were excluded first.
[0096] In order to further calculate the relative expression of each gene, the classification genes were further screened acco...
Embodiment 3
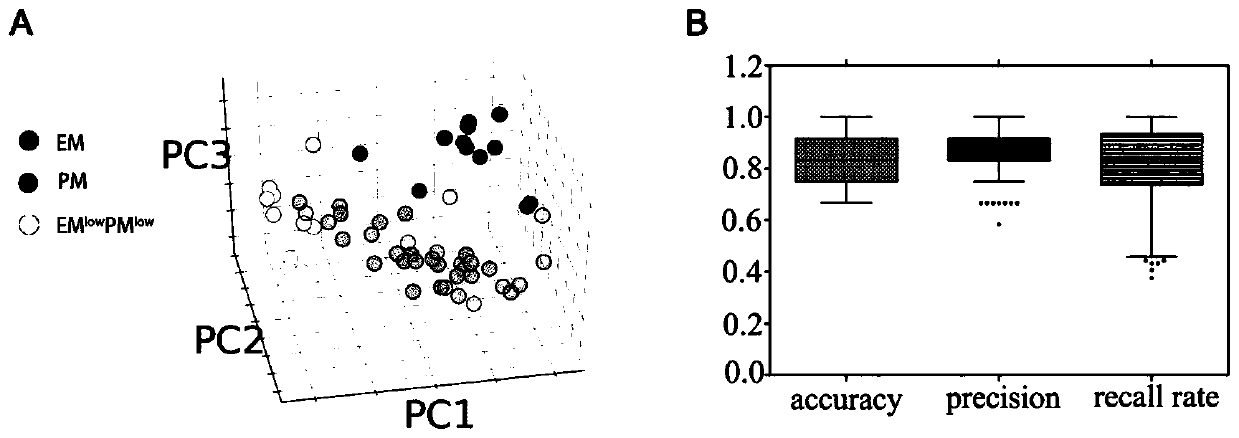
[0103] Example 3. Individualized molecular typing of gliomas using TLDA chips
[0104] 52 tumor samples were selected from the 263 samples described in Example 1, and 7 control normal brain tissue samples were added as the training sample set. Subsequently, the 48-gene Taqman low-density chip described in Example 2 was used to detect the gene expression profiles of these samples.
[0105] The detection method is as follows:
[0106] 1. Extract the total RNA of the sample and reverse transcribed it into cDNA.
[0107] 2. Use the Taqman low-density chip prepared in Example 2 to detect the expression levels of 44 typing genes, 3 internal reference genes, and quality control genes to be tested. The instrument used is the ABI 7900HT Fast real-time fluorescent quantitative PCR system. A 100μL reaction system was prepared, containing: 50μL 2×Taqman Universal PCR Master Mix with UNG (Thermo Fisher, catalog number 4352042), 48μL RNase-free water and 2μL template cDNA. Then the reaction syste...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com