Beta-isopropylmalate dehydrogenase and application of beta-isopropylmalate dehydrogenase in lipid synthesis
A technology of malate dehydrogenase and isopropyl, which is applied in the field of genetic engineering and microbial engineering, can solve problems such as difficult industrial production and limited capacity, and achieve the effect of promoting fatty acid production by microorganisms and improving biosynthesis ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1: Screening of genes encoding β-isopropylmalate dehydrogenase
[0053] Specific steps are as follows:
[0054] According to the sequence of the β-isopropylmalate dehydrogenase gene whose function has been identified in NCBI as a template, perform BLAST comparison in the gene bank of the sequenced M.alpinaATCC 32222 strain to obtain an alternative target gene; then The candidate genes were compared and screened for the second time in the NCBI library, and the final target gene was named MaLeuB (nucleotide sequence as shown in SEQ ID No.2), and the corresponding protein was named MaLeuB (amino acid sequence as shown in SEQ ID No. 2). shown in No.1).
[0055] The full length of cDNA corresponding to MaLeuB is 1146bp, encoding 381 amino acids. In order to further judge whether the screened MaLeuB belongs to isopropylmalate dehydrogenase, amino acid homology and conservative structure analysis were carried out between it and β-isopropylmalate dehydrogenase whose f...
Embodiment 2
[0057] Example 2: Cloning of MaLeuB
[0058] Specific steps are as follows:
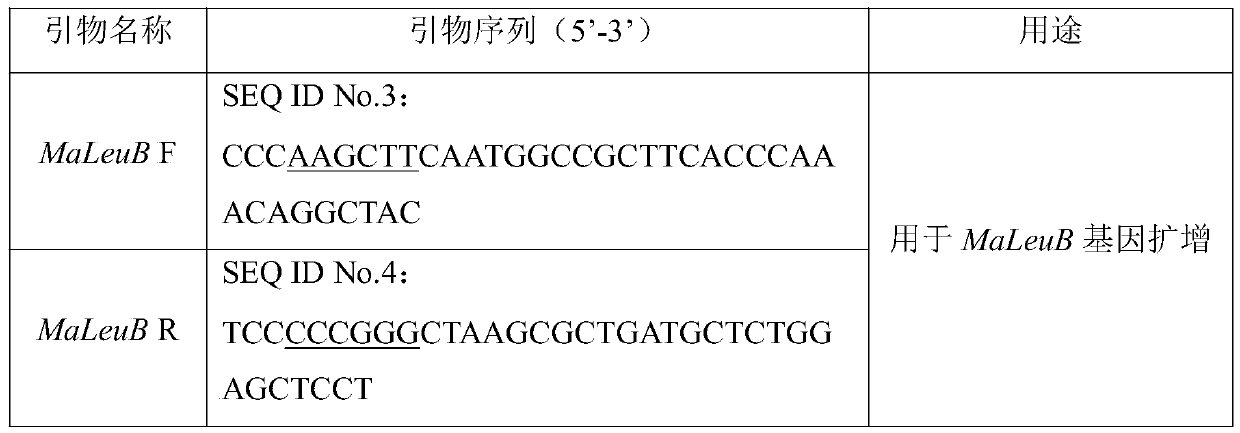
[0059] The total RNA of Mortierella alpina (Mortierella alpina) ATCC 32222 was extracted using the Trizol method, and cDNA was obtained by reverse transcription according to the instructions of the Takara reverse transcription kit. The reaction amplifies MaLeuB, and the primers used to amplify MaLeuB are listed in Table 1.
[0060] The PCR instrument used is BIO-RAD T100 Thermal Cycler, using KOD plus high-fidelity DNA polymerase, the reaction system is 50 μL, and the content of the system is carried out according to the instructions of the DNA polymerase; the reaction process is as follows: pre-denaturation at 95°C for 5 minutes, then denaturation at 95°C 30s, annealing at 55°C for 30s, extension at 68°C for 1.5min, repeat the above three steps 30 times, then fully extend at 68°C for 5min, and finally drop to 12°C for 10min and stop.
[0061] After the reaction was completed, the amplified product...
Embodiment 3
[0064] Example 3: Expression of MaLeuB in Mortierella alpina
[0065] Specific steps are as follows:
[0066] (1) Construction of Mortierella alpina expression vector
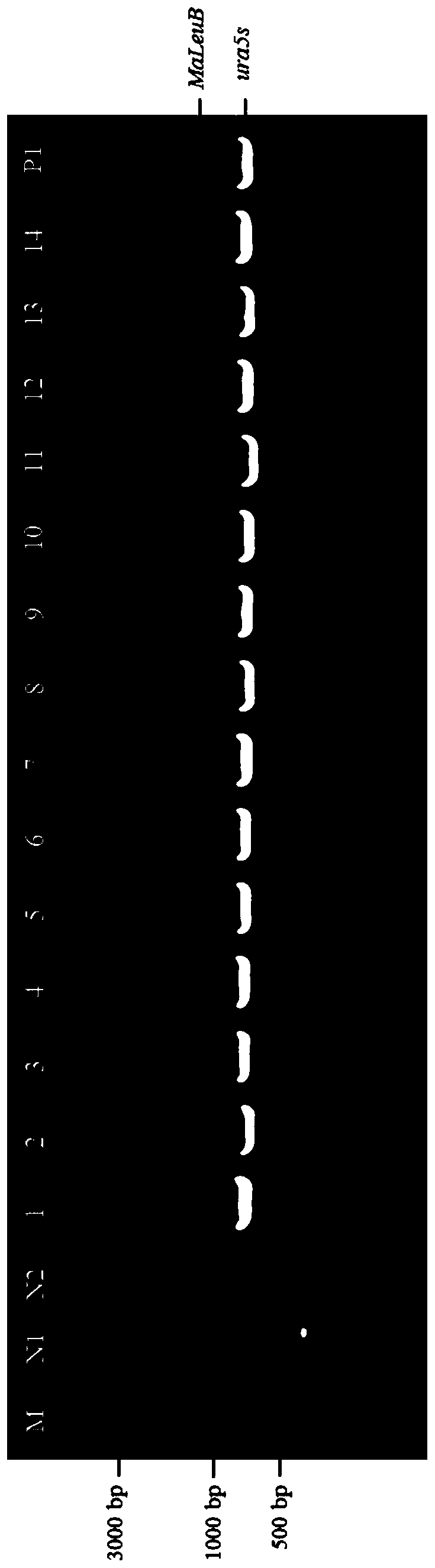
[0067] MaLeuB obtained in Example 2 and the expression vector pBIG2-ura5s-ITs were digested with restriction endonucleases HindⅢ and SmaI, and then T 4 Ligase ligated the digested and purified DNA to obtain a ligation product. The specific enzyme digestion system (20 μL) is shown in Table 2.
[0068] Table 2 enzyme digestion system
[0069] Reagent Dosage 10×cutmart buffer 2μL restriction endonuclease 1μL PCR product or vector 200ng~1μg wxya 2 o
Make up to 20μL
[0070] After connecting the obtained ligation product overnight at 4-16°C, transform it into Escherichia coli DH5α competent cells. The transformation method is as follows: take 100 μL of competent cells in a sterile state, add 5-8 μL of the ligation product, and mix by pipetting; Transfer the mixed c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com