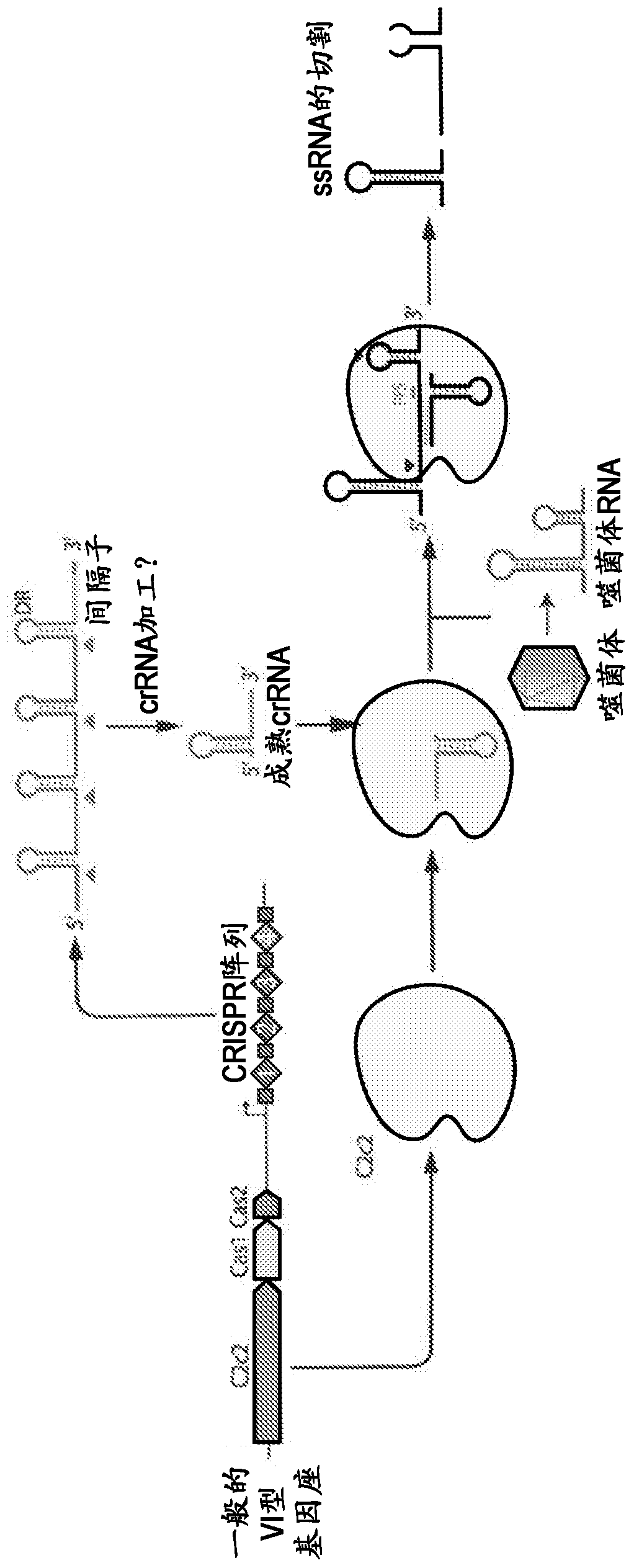
Crispr effector system based diagnostics
An effector protein, detection system technology, applied in ICT adaptation, genetic engineering, microbial assay/inspection, etc., can solve problems such as limited operator availability, dependence, low sensitivity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 2
[0547] Example 2 - Highly sensitive and specific detection of C2C2-mediated DNA and RNA from LEPTOTRICHIA WADEI
[0548] Rapid, cheap, and sensitive nucleic acid tests could facilitate pathogen detection, genotyping, and disease surveillance at the point of care. The RNA-guided, RNA-targeting CRISPR effector Cas13a (formerly known as C2c2) exhibits a "collateral effect" of promiscuous RNase activity upon target recognition. Applicants combined the parallel effect of Cas13a with isothermal amplification to create a CRISPR-based diagnostic (CRISPR-Dx), providing rapid DNA or RNA detection with attomolar sensitivity and single-base mismatch specificity. Applicants used this Cas13a-based molecular detection platform, called SHERLOCK (Specific High Sensitivity Enzyme Reporter Unlocked), to detect specific strains of Zika virus and dengue virus, differentiate pathogenic bacteria, genotype human DNA, and identify Cell tumor DNA mutation. In addition, SHERLOCK reaction reagents can ...
Embodiment 3
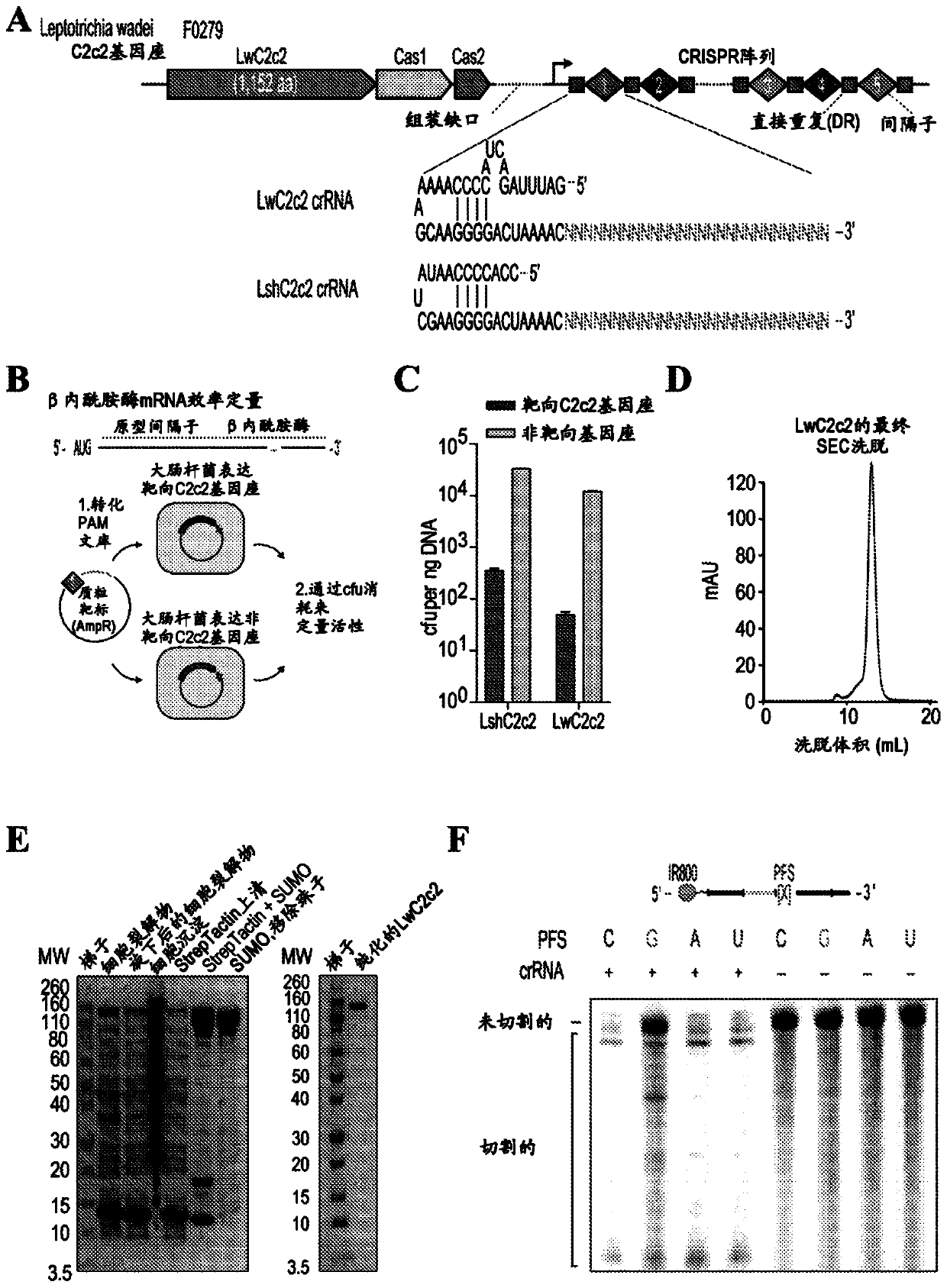
[0660] Example 3 Characterization of Cas13b Orthologs with Orthogonal Base Preferences
[0661] Applicants biochemically characterized 14 orthologs of the recently defined Type VI CRISPR-Cas13b family of RNA-guided RNA-targeting enzymes to find new candidates for improved SHERLOCK assay technology ( Figure 83 A and 85). Applicants were able to heterologously express 14 Cas13b orthologs in E. coli and purify the proteins for in vitro RNase activity assays ( Figure 86 ). Because different Cas13 orthologs may have different base preferences for optimal cleavage activity, Applicants generated a fluorescent RNase homopolymer sensor composed of 5 A, G, C, or U to assess orthogonal cleavage preference. Applicants incubated each ortholog with its cognate crRNA targeting the synthetic 173nt ssRNA 1 and measured parallel cleavage activity using a homopolymer fluorescent sensor ( Figure 83 B and 87).
Embodiment 4
[0662] Example 4 Motif Discovery Screening Using Libraries
[0663] To further explore the diversity of cleavage preferences of various Cas13a and Cas13b orthologs, Applicants developed a library-based approach to characterize motifs of endonuclease activity preference in response to parallel activity. Applicants used a degenerate 6-mer RNA reporter flanked by a constant DNA handle, which allows amplification and readout of the uncut sequence ( Figure 83 C). Incubation of the library with parallel activated Cas13 enzymes resulted in detectable cleavage and was dependent on the addition of the target RNA ( Figure 88 ). Sequencing of the depleted motif revealed an increase in the skewness of the library over the digestion time ( Figure 89 A), base preference is indicated, and selection of sequences above a threshold ratio yields enriched sequences corresponding to cleavage by the enzyme ( Figure 89 B). Sequence logos from enriched motifs reproduced the U preference obse...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com