Methods for engineering host cell genomes and uses thereof
A host cell and gene technology, applied in the fields of cell biology, bioengineering, and molecular biology, can solve the problems of difficult screening, increased uncontrollable uncertainty of cell lines, and heterogeneity of cell lines
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
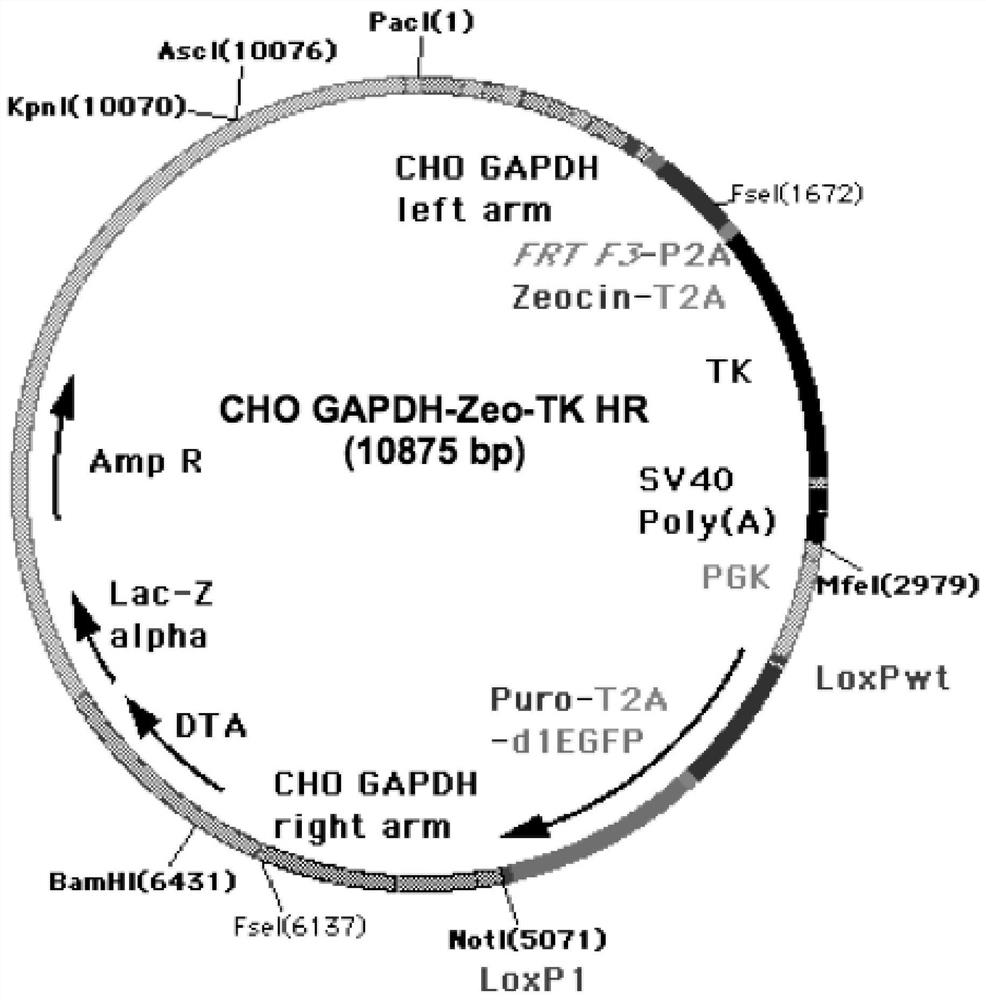
[0208] Example 1: Construction of CHO cell targeting vector and establishment of anchor cell line
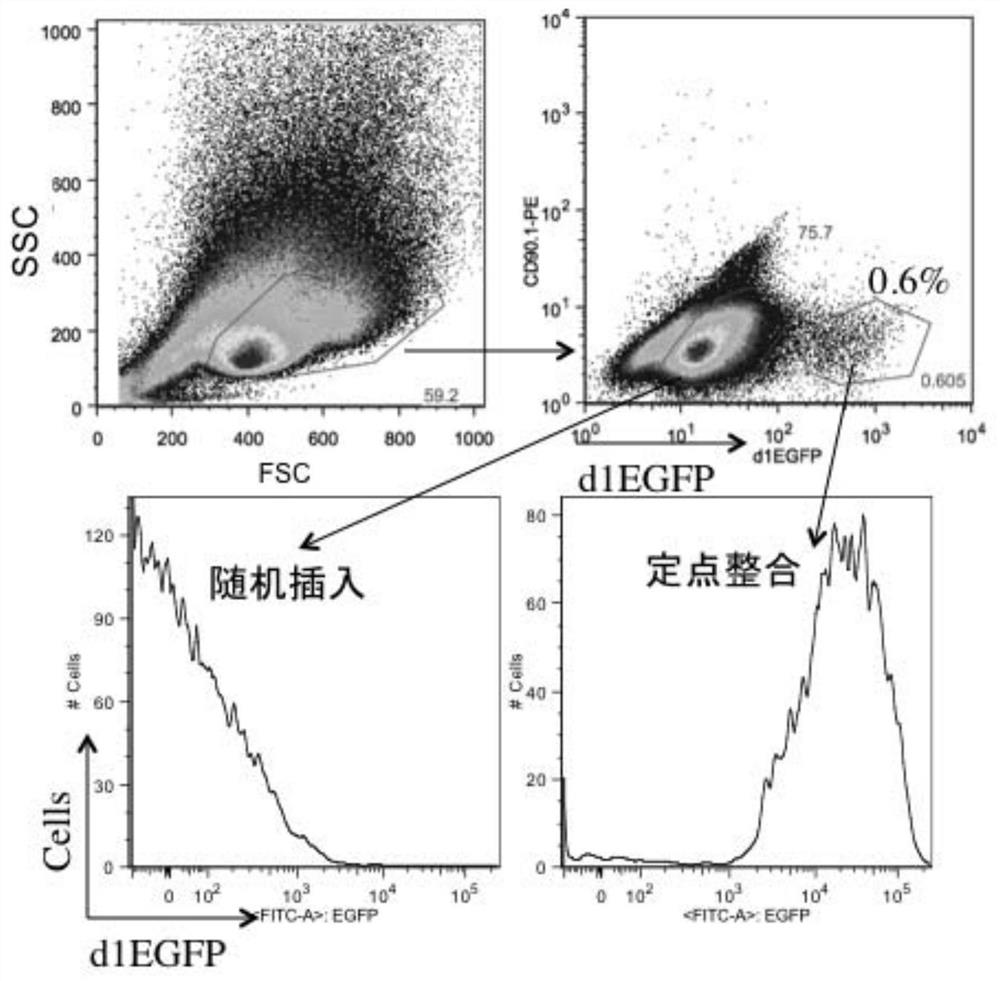
[0209] In order to achieve continuous, cyclical, and unlimited site-directed integration in the CHO cell genome, it is first necessary to find a high-expression hot spot (Hot-Spot) in the CHO cell genome. After in-depth research, the inventor found that the promoter (Promoter) of the highly constitutively expressed housekeeping gene GAPDH meets the requirements of open chromosome structure and constant high expression. For example, Bahr S. et al. identified a site similar to mouse ROSA26 in CHO cells. ROSA26 is a known structurally open and constant high expression site in the mouse genome, and is often used to implant foreign genes to prepare transgenic mice. Although the ROSA26 gene also exists in CHO cells, the transcriptional intensity of its mRNA level is lower than that of several other invariant housekeeping genes, especially only 1 / 300 of the transcriptional intensi...
Embodiment 2
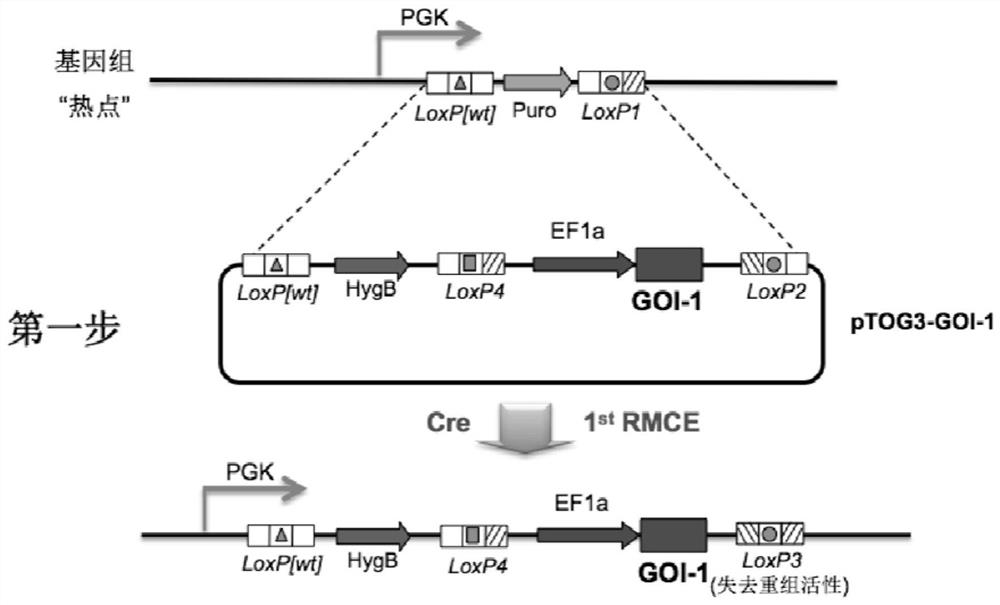
[0215] Example 2: Construction of "Toggle-In" vector and method for stable co-expression of multiple genes
[0216] In order to solve the problem of stable co-expression of multiple genes in the transformation of CHO cell lines, the inventors developed a set of "Toggle-In" platform for Cre-LoxP-mediated homologous recombination based on the aforementioned anchored CHO cell lines. system. The feature of this system is that it can achieve continuous, cyclical, and unlimited genome site-directed integration with two vectors (pTOG3 and pTOG4) in sequence and only two drug resistance genes (hygromycin B and puromycin). , these CHO cell clones, each of which is isogeneic, can achieve 1:1 expression of the insert.
[0217] pTOG3 (its nucleotide sequence is shown in SEQ ID NO: 11, which carries the DsRed-E2 tracer protein, and its nucleotide sequence is shown in SEQ ID NO: 36) and pTOG4 (its nucleotide sequence is shown in SEQ ID NO: 12, which carries the d1EGFP tracer protein, an...
Embodiment 3
[0229] Example 3: Knockout of Fut8 gene in CHO cells using TALEN technology
[0230] In order to conveniently and quickly knock out the Fut8 gene in CHO cells, the inventors constructed a pair of TALEN vectors targeting the 10th exon of the CHO cell Fut8 gene (the DNA sequence encoding the left arm of the TALEN protein for knocking out the Fut8 gene in CHO cells is shown in SEQ ID NO. : 18, the DNA sequences encoding the right arm of the TALEN protein that knocked out the Fut8 gene in CHO cells are shown in SEQ ID NO: 19), named FL#2 and FR#12, respectively. 5 μg of the plasmids prepared by the two miniPrep were taken and introduced into 1 million CHO-K1 cells (ATCC, CCL-61) by electroporation. The electroporator is the Neon Electroporation system from LifeTech. Electric transfer conditions are 1620v, 10ms, 3 pulses. After electroporation, CHO cells were cultured in DMEM medium containing 5% FBS for 8 days without any drug selection. This growth stage allows those clones ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com