Capture probe composition of three subtypes of thalassemia and its application method and application device
A capture probe and anemia technology, which is applied in the field of capture probe compositions of three subtypes of thalassemia, can solve the problems of low detection scheme and no cost comparison, and achieve the effect of easy detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
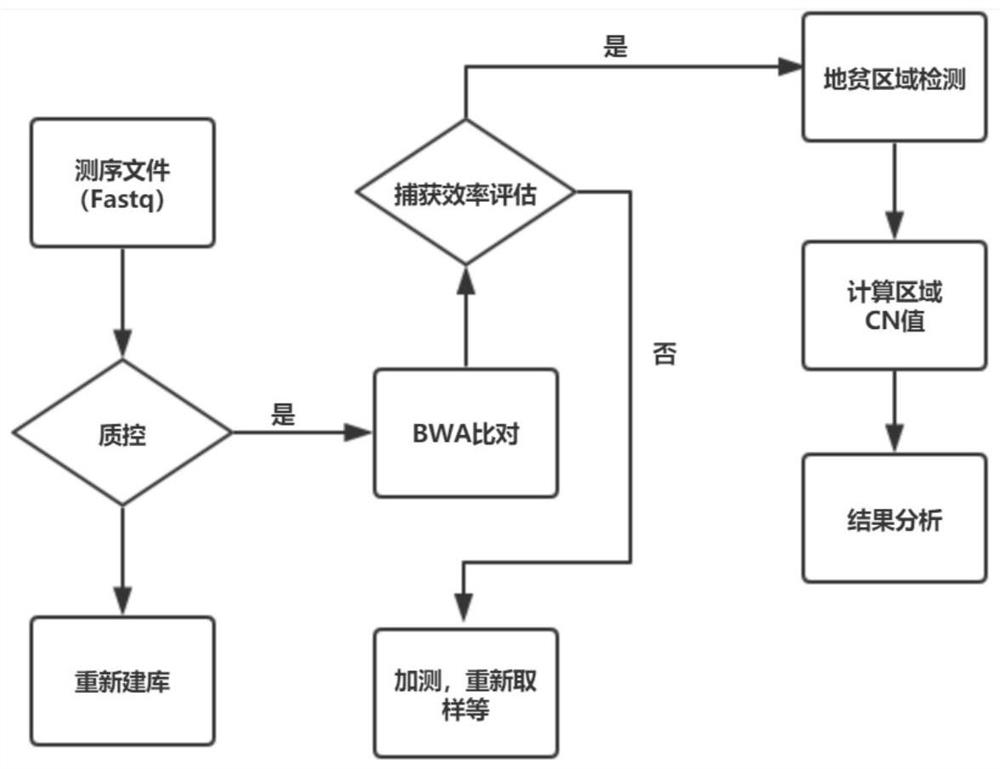
[0032] Based on the above research results, in a typical embodiment of the present application, a method for preparing a capture probe composition of the three subtypes of thalassemia is provided, the preparation method comprising: combining the three subtypes of thalassemia The full-length region of the relevant gene is divided into different small fragments according to the preset length (for example, 120bp, 150bp or other suitable length); compare each small fragment to the human reference genome to obtain the first single alignment Region set and multi-comparison region set; the multi-comparison region set is moved forward or backward by kbp on the human reference genome, k≤100, and the new region after the shift is compared with the human reference genome again to obtain the first Two sets of single alignment regions; performing probe design from the first set of single alignment regions and the second set of single alignment regions to obtain a capture probe composition. ...
Embodiment 1
[0059] Embodiment 1 capture probe design
[0060] First, select the full-length regions of the three types of thalassemia genes, SEA deletion, -α4.2 deletion and -α3.7 deletion, and separate them into different small fragments according to the length of 120bp, and compare each small fragment to human by blast On the hg19 reference genome, some unique alignment regions and some multiple alignment regions were obtained.
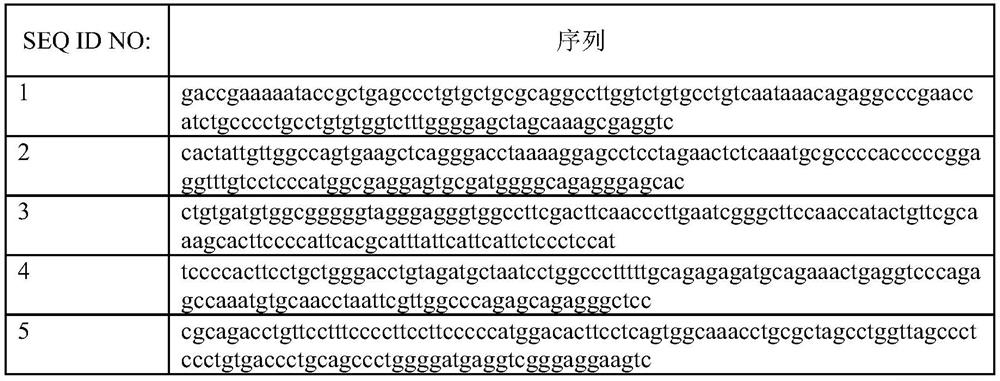
[0061] Make adjustments for multiple alignment regions. The adjustment method is to move these regions forward or backward on the genome by less than 100 bp, compare the moved new region with hg19, and finally select the region of the unique comparison result generated by the above two steps for probe design. The specific probe design rules are: 1) The probe must have only one alignment result in the deletion region; 2) Evenly cover three different deletion regions. Finally, 71 uniquely aligned probes (as shown in Table 1) were finally determined as the final...
Embodiment 2
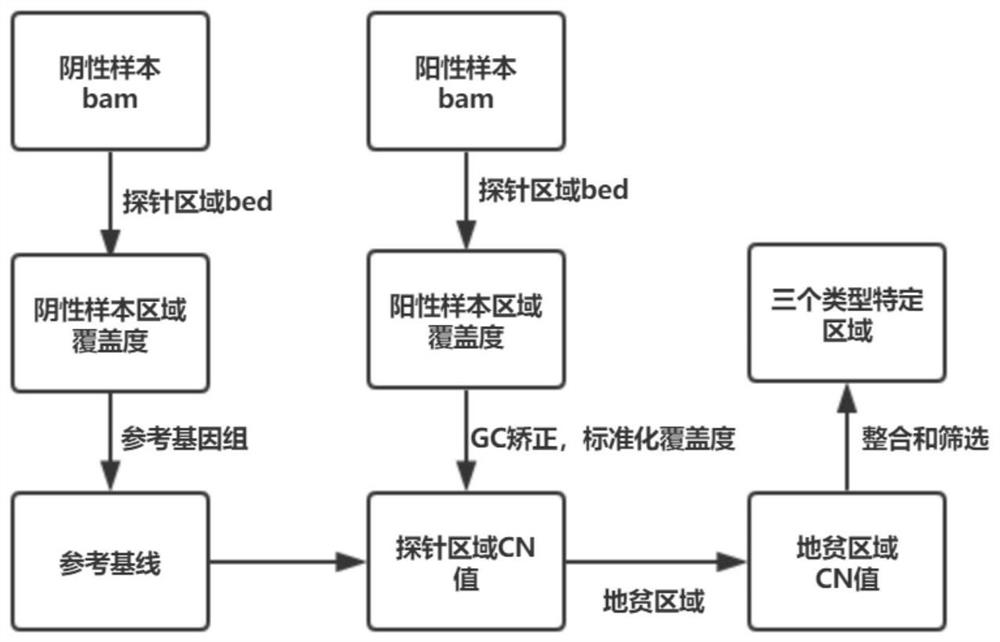
[0062] Example 2 Using a negative sample set to establish a reference baseline for coverage depth of negative samples
[0063] DNA extraction was performed on the 25 negative samples collected to build a library, the target region was captured, and then sequenced on the machine.
[0064] The raw data (raw Data, stored in the FASTQ format) obtained by removing adapters and index tags from the sequencing data, filtered out low-quality data through quality control, and obtained effective data (clean data).
[0065] Align the effective data (clean data) to the human reference genome hg19 through general-purpose alignment software (such as BWA), and obtain the bam file of the alignment result.
[0066] Based on the existing negative samples, construct a baseline for subsequent determination of whether there is a copy number deletion: use the above comparison to obtain the bam file of the result, and use the bed file of the capture probe region to calculate the coverage depth of the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com