Method for predicting tumor neoantigen and its application
A prediction method and tumor technology, applied in the field of biomedicine, can solve the problems that tumor vaccines cannot meet the needs of non-response to treatment and produce drug-resistant patients, and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
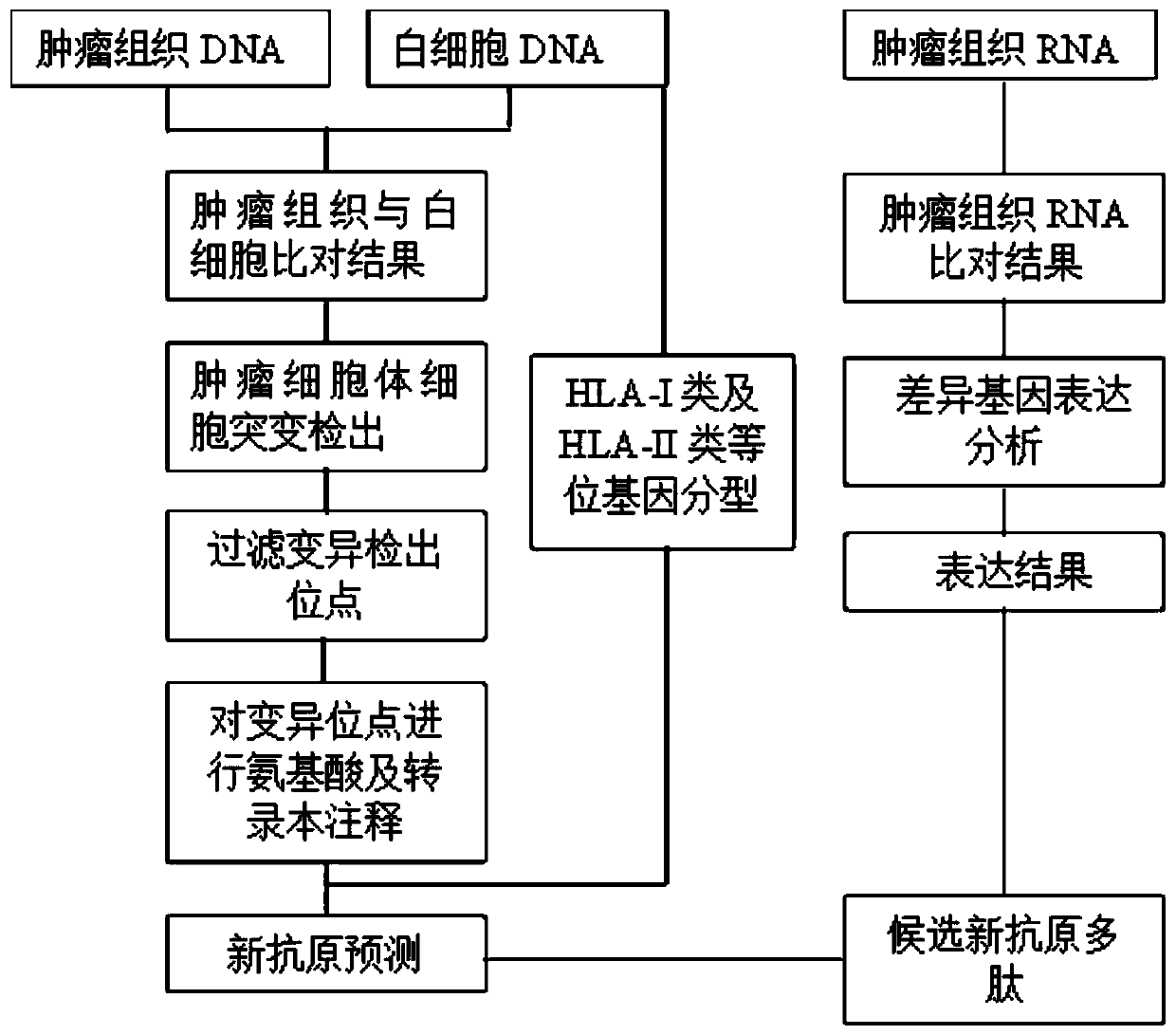
[0057] In this example, the whole exome sequencing and RNA-Seq data of melanocytoma patients were used for testing, which can accurately type HLA-I and HLA-II alleles, and predict that they can be compared with HLA-I Class and HLA-II allele tightly bound neoantigens.
[0058] Specific steps are as follows:
[0059] The tumor tissue and plasma white blood cell samples of a melanoma patient were obtained, and the tumor tissue DNA, RNA and plasma white blood cell DNA were extracted to construct a sequencing library, and whole exome sequencing was performed to obtain sequencing data.
[0060] The data analysis is as follows:
[0061] step 1:
[0062] 1) Plasma leukocyte DNA comparison: Use Fastqc (v 0.11.6) (-t 6) software to count the sequencing quality of plasma leukocyte DNA, and use Cutadapter (v1.2.1) software to remove the sequencing adapters connected when building the library. The software uses the default Parameters, remove sequences whose bases with a quality score le...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com