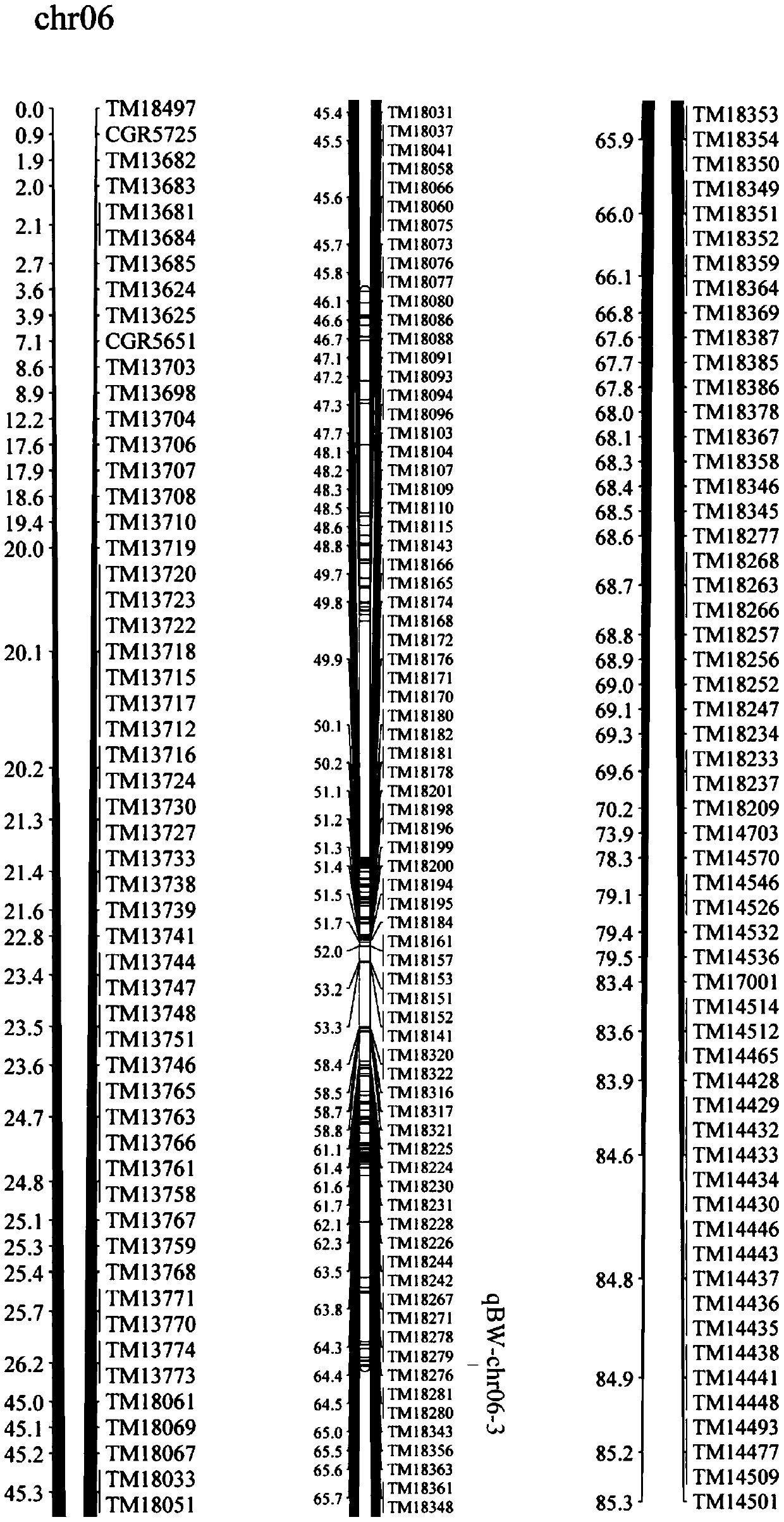
SNP molecular marker derived from single-boll weight major gene of Xinluzao 24
A major gene and molecular marker technology, applied in the field of biotechnology applications, can solve the problems of large QTL interval, affecting application, and inability to detect across groups
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0028] The present invention will be further clarified below by the detailed description of the specific embodiment, and the molecular marker linked with the main effect gene of upland cotton boll weight is obtained by the following method:
[0029] (1) The breeding method and process of the recombinant inbred line used to identify the boll weight of upland cotton and to construct the genetic linkage map: the upland cotton cultivar Lumianyan 28 with high-yielding potential cultivated by Shandong Cotton Research Center As the female parent, Xinluzao 24, a high-quality variety cultivated by Xinjiang Kangdi Company, was used as the male parent. In the summer of 2008, the hybrid combination was configured in the experimental farm of the Cotton Research Institute of the Chinese Academy of Agricultural Sciences (Anyang, Henan). At the end of 2008, F was planted in Hainan. 1 . Planted 238 F in Anyang in the spring of 2009 2 Single plant, obtained F after selfing 2 : 3 families. Pi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com