Method for increasing expression of survival motor neuron (SMN) protein based on gene editing technology, and application of method in spinal muscular atrophy (SMA) treatment
A protein expression and gene technology, applied in the field of genetic engineering, can solve the problems of high price, unacceptable for patients, and short-term functional SMN protein.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0104] Example 1, CRISPR / Cas9-mediated gene editing
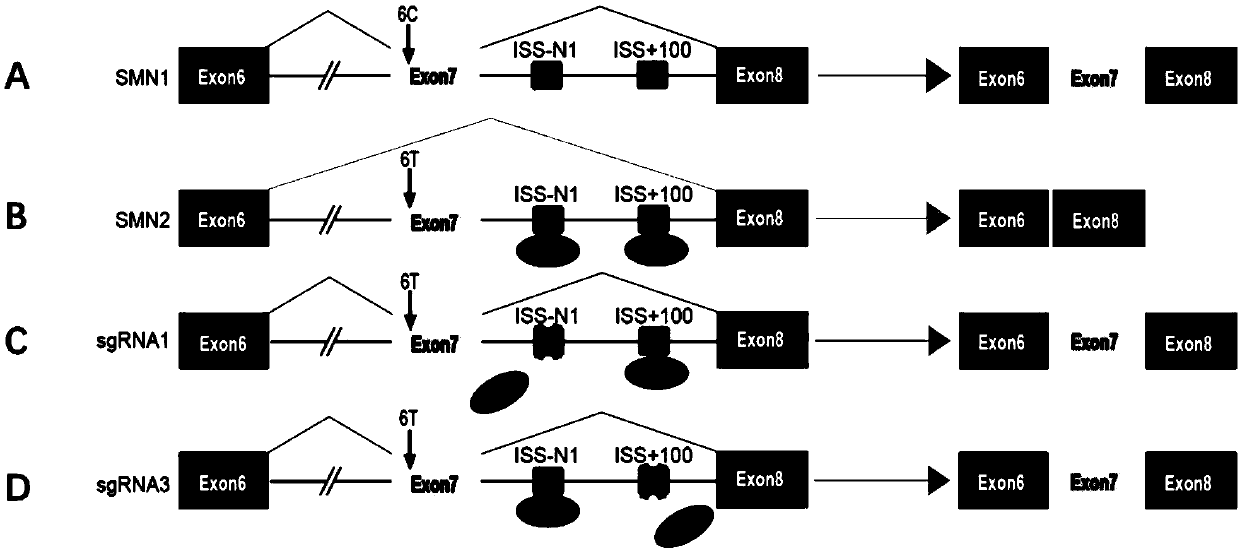
[0105] SMN2 is a highly similar gene to SMN1, but due to the difference in C6T on exon 7 of SMN2, exon 7 is skipped during the splicing process, such as figure 1 A ~ B shown. The intron splicing silencers ISS-N1 and ISS+100 are located in exon 7. In order to prevent this exon skipping, the inventors used CRISPR / Cas9 to target ISS-N1 or ISS+100 in an attempt to destroy their structure. The gene splicing situation and the CRISPR / Cas9 editing strategy of the present invention are as follows: figure 1 shown.
[0106] In order to disrupt ISS-N1 or ISS+100, the inventors designed sgRNA at a position adjacent to or inside ISS-N1 or ISS+100. According to the position difference, the present inventors prepared a large number of sgRNAs to screen suitable sgRNAs, and found gRNAs suitable for destroying ISS-N1 or ISS+100 after repeated research and screening. The sequence of the sgRNA is as follows:
[0107] sgRNAs used to target...
Embodiment 2
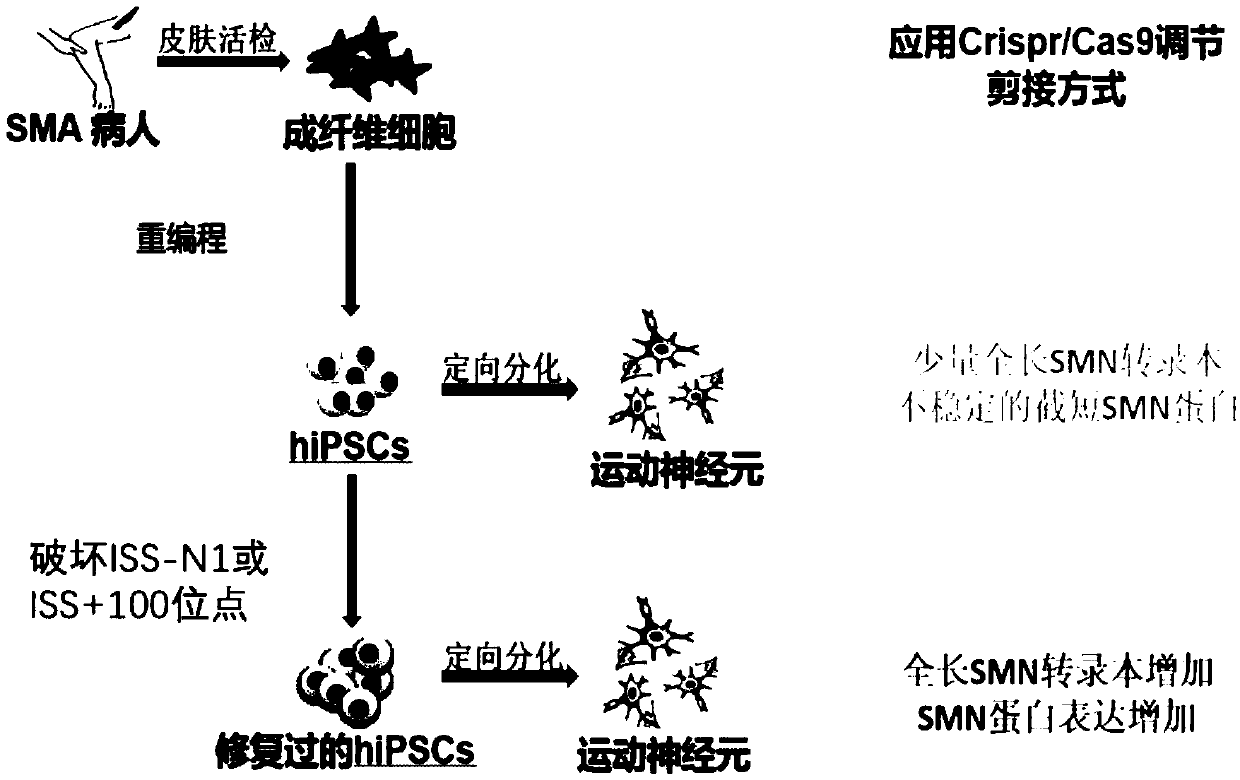
[0115] Example 2. The splicing pattern of iPSC cell line SC-SMA that destroys ISS-N1 or ISS+100 is regulated and changed
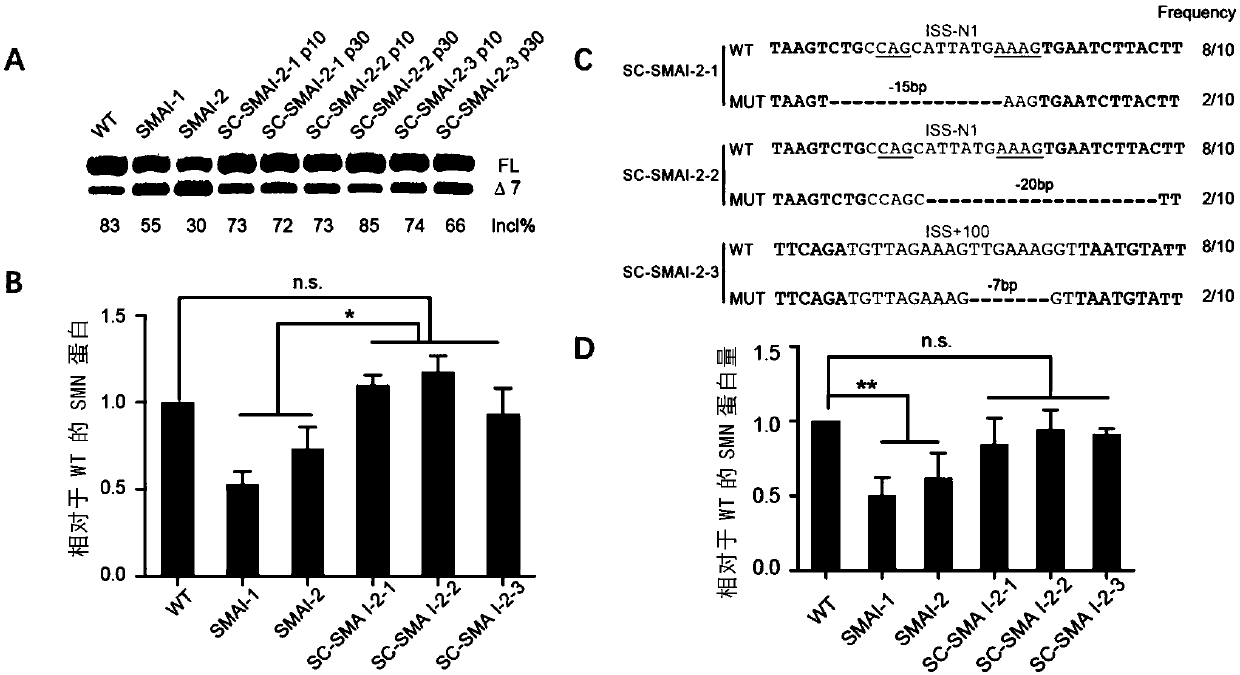
[0116] In the monoclonal cell lines obtained in Example 1, the inventors detected two ISS-N1 disrupted cell lines, which were named SC-SMAI-2-1 (gene editing guided by sgRNA1), and SC-SMAI-2 -2 (gene editing guided by sgRNA1); and, an ISS+100 disrupted SC-SMAI-2-3 cell line (gene editing guided by sgRNA3) and mycoplasma-free. The present inventors performed RT-PCR analysis and qPCR analysis to determine the SMN2 gene splicing and SMN protein expression after destroying ISS-N1 or ISS+100 at the cellular level.
[0117] see results image 3 . As a result of RT-PCR analysis, the promotion of the full-length SMN2 (SMN-FL) transcript can be significantly seen ( image 3 , A). Moreover, the results of Western blot showed that the SMN protein was 1.7 times that of unedited ( image 3 , B).
Embodiment 3
[0118] Example 3, CRISPR / Cas9-mediated gene editing can prevent the occurrence of SMA
[0119] 1. Obtain mice with significant disruption of ISS-N1
[0120] Type III mice contain 4 copies of the SMN2 gene and no mouse Smn gene (Smn - / - ; SMN2 tg / tg ). Type III mice do not develop the SMA phenotype, but develop necrosis of the ears and shortening of the tail within a month or so.
[0121] The inventors used SpCas9, SaCas9mRNA and their corresponding sgRNA (sgRNA1, sgRNA2) to inject type III mice (Smn - / - ;SMN2 tg / tg ) and BH heterozygous mice (Smn - / + ) Fertilized eggs obtained from cages ( Figure 4 , A). As a result, there is a 50% chance of obtaining type I SMA mice (Smn - / - ;SMN2 tg / - ) and 50% likely to get heterozygous mice (Smn + / - ;SMN2 tg / - )( Figure 4 , A).
[0122] The average survival days of SMA mice was 12 days (n=35). The present inventors applied CRISPR / Cas9 gene editing technology to interfere with the splicing of SMN2 at the level of fertilized ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com