Joint composition and application thereof
A composition and linker connection technology, applied in combinatorial chemistry, organic compound libraries, biochemical equipment and methods, etc., can solve problems such as inability to remove false positives, and achieve the goal of improving utilization efficiency, increasing library yield, and improving utilization. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
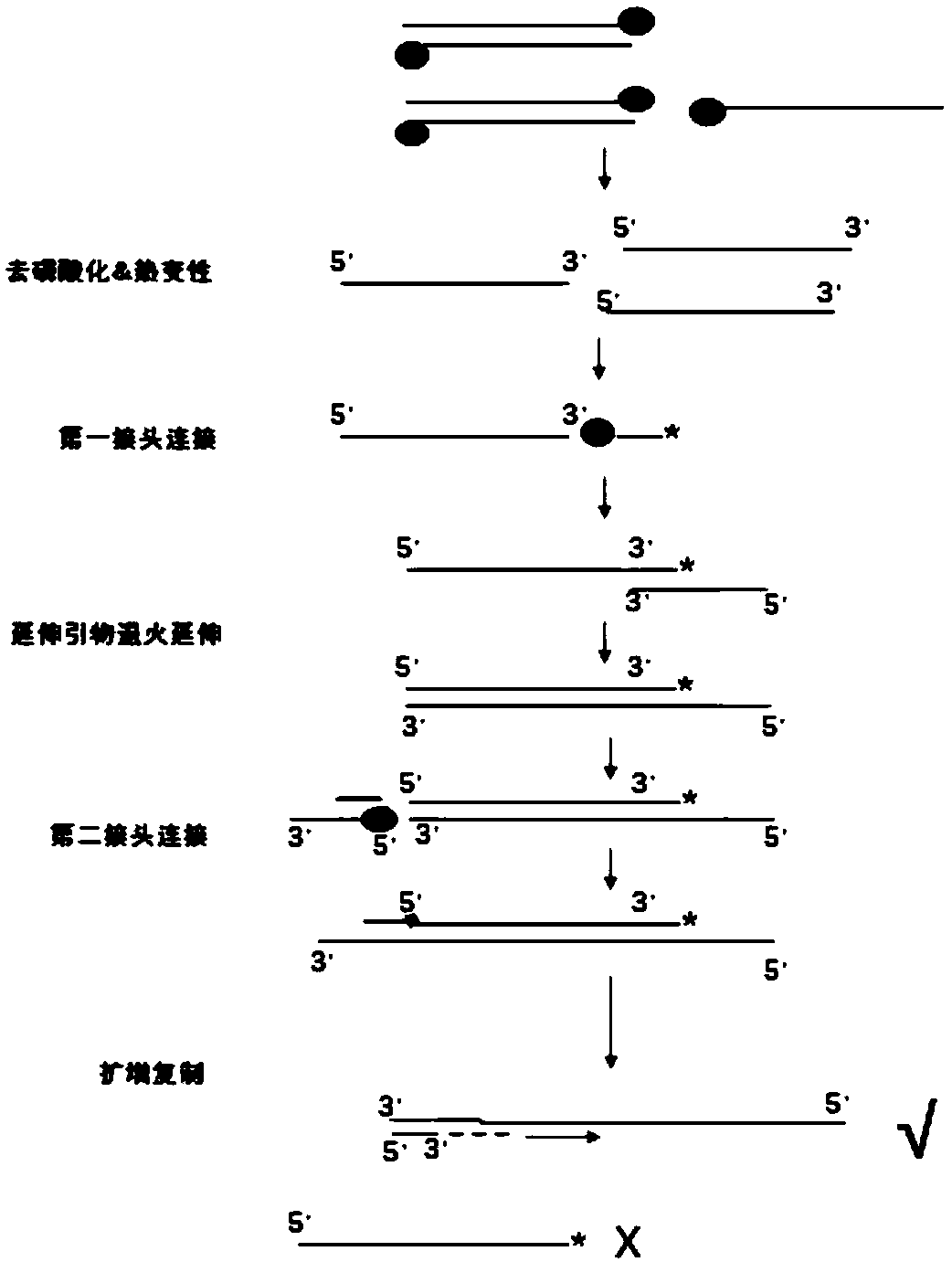
[0070] Example 1 BGI-seq500 platform cfDNA starting single-stranded UID library construction
[0071] Based on BGI-seq500 platform of BGI, this example provides a cfDNA starting single-stranded UID library construction method, in which cfDNA is derived from cfDNA extracted from mixed plasma, and the mixed plasma is derived from BGI Gene Cell Resource Bank, including the following specific steps :
[0072] 1) 3ng cfDNA was dephosphorylated at the 5' end, denatured and melted, and DNA repaired. The specific system preparation process is shown in Table 2 below:
[0073] Table 2
[0074] system volume / μl dna 1-100ng (volume X<=16μl)
Circular Ligase Buffer II 4 MnCl 2
2 Endonuclease VIII (10U / μl) 0.5 nuclease free water 16-X
[0075] Incubate at 37°C for 1 hour; then add 2 μl FastAP (1U / μl), incubate at 37°C for 10 minutes, at 95°C for 2 minutes, and incubate on ice for 2 minutes;
[0076] 2) Connect the first linker to the si...
Embodiment 2
[0103] Gradient optimization of the first joint length in embodiment 2
[0104] Since it was found that the length of the first linker had a greater impact on the ligation efficiency, the choice of linker was due to the fact that the first linker consisted of a 5bp UID sequence + another sequence, which was the reverse sequence truncated from the 3' end of the extension primer. To the complementary sequence, and since the first adapter needs to be blocked by ddc, the end is required to be base C, and the extension primer should be G, that is, this embodiment uses truncated 20bp, 24bp and 34bp adapters for library construction, so that Analyze and study the influence of linkers with different lengths on the connection efficiency and the final library construction effect. The specific steps of the library construction method are the same as in Example 1. The specific sequences and results of the linkers are shown in Table 11 below:
[0105] Table 11
[0106]
[0107]
[0...
Embodiment 3
[0126] Example 3 FFPE severely degraded sample initial single-chain UID library construction
[0127] Since the present application can also build a library for degraded DNA, this embodiment provides a method for building a library of starting single-stranded UID from FFPE severely degraded samples, including the following specific steps:
[0128] 1) Extract FFPE sample DNA;
[0129] 2) Covaris ultrasound interrupts and enriches DNA samples, 12 cycles;
[0130] 3) Purification with 1.5×Agencourt AMPure XP-Medium magnetic beads;
[0131] 4) Wash twice with 80% alcohol, redissolve in 20 μl;
[0132] 5) Qubit quantification;
[0133] 6) Take 30ng of interrupted FFPE genomic DNA, and build a library according to the method of building a library in Implementation 1;
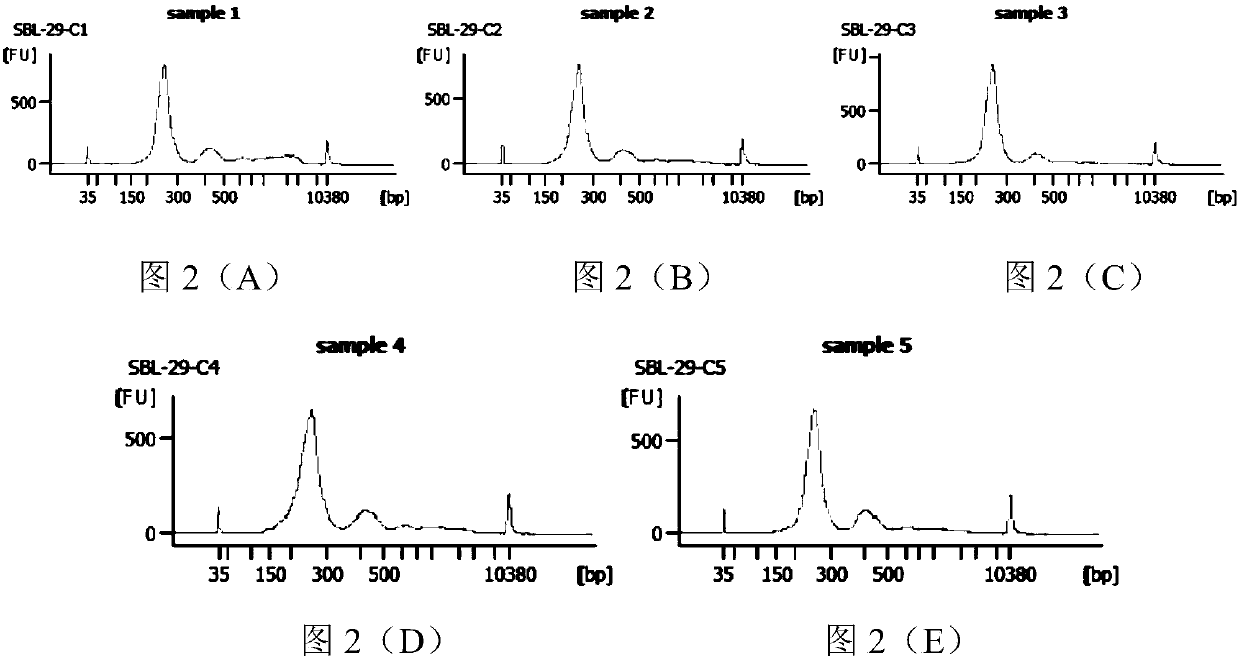
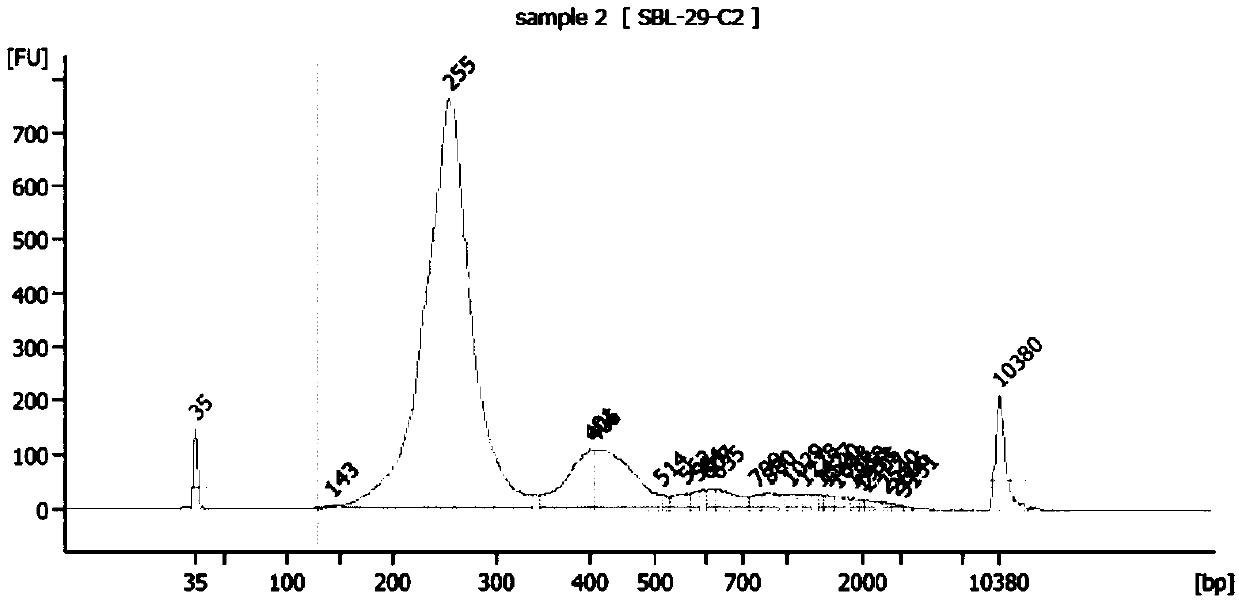
[0134] 7) Qubit quantification and Agilent 2100 detection, the results are as follows Figure 6 (A)- Figure 6 (C) and Figure 7 shown.
[0135] From Figure 6 (A)- Figure 6 (C) It can be seen that the dist...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com