Nucleotide sequence and vector for directional cloning
A nucleotide sequence and carrier technology, applied in the field of molecular biology, can solve problems such as low technical requirements, high cost, and high enzyme cutting conditions, and achieve the effects of ensuring positive rate, preventing self-ligation, and reducing costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
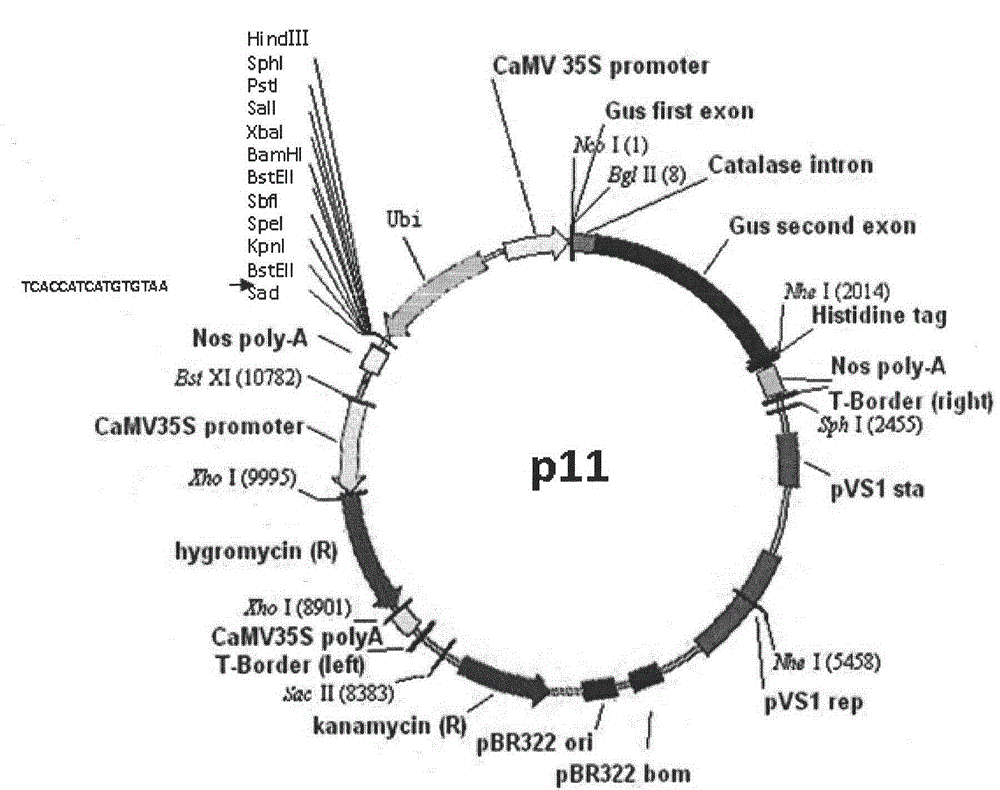
[0047] Example 1 P11 vector construction
[0048] 1. PCR amplification of maize Ubiquitin promoter fragment and construction of pMD18-T+Ubi recombinant vector
[0049] Ubi promoter PCR amplification
[0050] Genomic DNA of maize variety B73 (Zea mays mays cv. http: / / www.maizegdb.org / ), according to the sequence of Ubi promoter in maize B73 gDNA, a pair of PCR-specific amplification primers were designed at the beginning and end respectively (upstream primer F1: GG CTGCAG TGCAGCGTGACCCGGTCGT (SEQ ID NO: 1), plus restriction site PstI and protection base, downstream primer R1: GG CTGCAG AAGTAACACCAAAC (SEQ ID NO: 2), plus restriction site Pst I and protection bases). Using the gDNA of corn B73 extracted above as a template, high-fidelity Ex Taq TM (TaKaRa, DRR100B) polymerase for PCR amplification. As shown in Table 1.
[0051] Table 1 PCR system for gene promoter amplification
[0052] Composition Volume (μl)
[0053] Genomic DNA 0.2
[0054] dNTPs (2.5mM) 2
[...
Embodiment 2
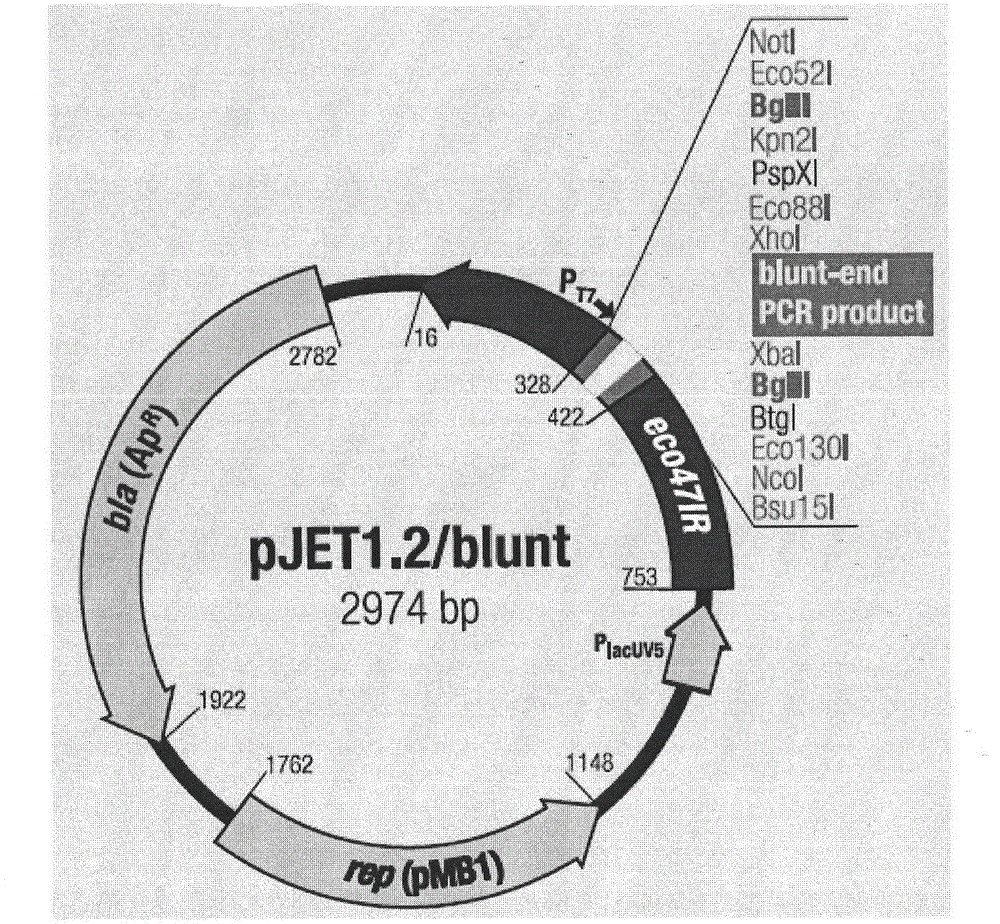
[0144] The PCR amplification of the target gene CaMV35S promoter sequence fragment of embodiment 2 (Primestar amplification Increase) and the construction of the recombinant vector DJET+A
[0145] 1. PCR amplification
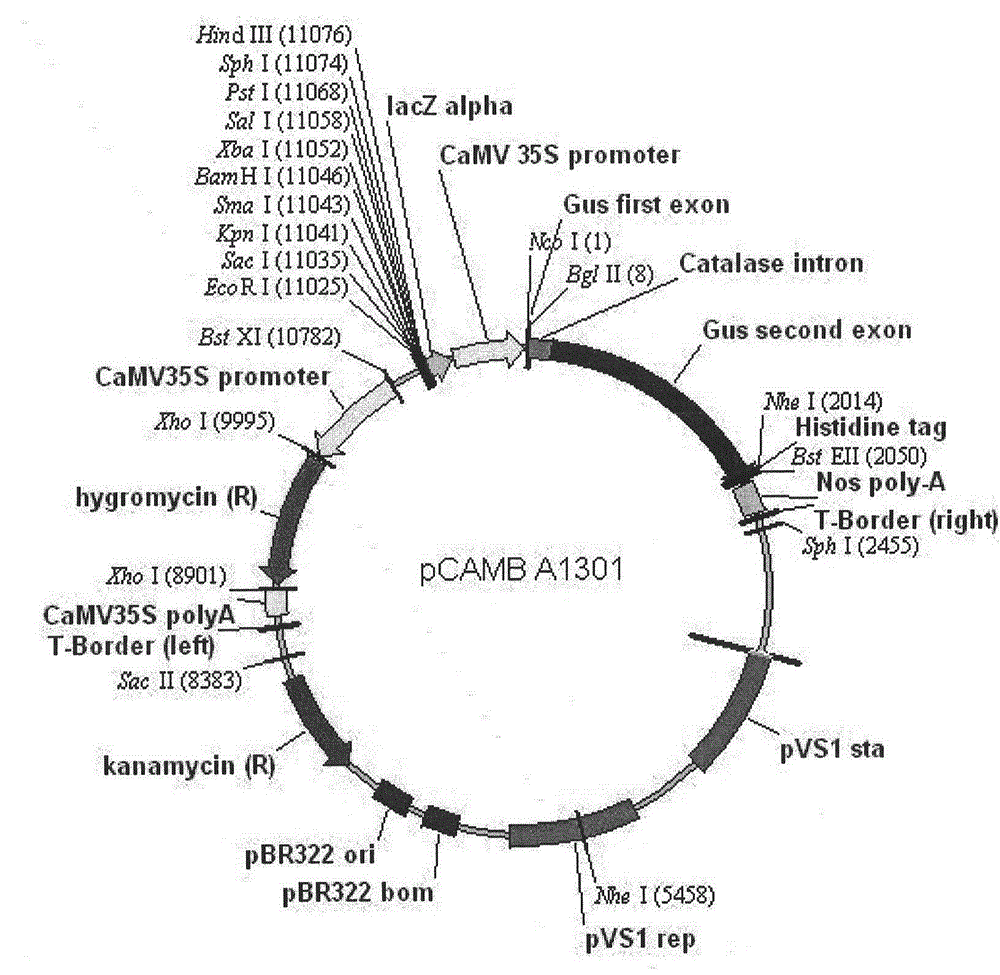
[0146] According to the CaMV35S promoter sequence in the pCAMBIA-1301 plasmid, a pair of gene-specific primers (upstream primer 35S-F, plus restriction enzyme site BstE Ⅱ (GGT TACC) and protective base were designed at the beginning and end respectively, and downstream primer 35S- R, plus restriction enzyme cutting site BstE Ⅱ (GGT GACC) and protective bases), using pCAMBIA-1301 plasmid as a template, high-fidelity Taq polymerase for PCR amplification.
[0147] 35S-F: A GGTTACC GCTTCATGGAGTCAAAGATTC (SEQ ID NO: 9)
[0148] 35S-R: A GGTGACC TCTACCATGGTCAAGAGTCC (SEQ ID NO: 10)
[0149] Gene amplification PCR system: 25μl
[0150] wxya 2 O 10.3 μl
[0151] 2×Primer Star Mix 12.5μl
[0152] pCAMBIA-1301 plasmid 0.2μl
[0153] Primer 35S-F 1μl
[01...
Embodiment 3
[0168] The PCR amplification (Primestar amplification) of the target gene hygromycin resistance gene sequence fragment of embodiment 3 Increase) and construction of recombinant vector pJET+B
[0169] 1. PCR amplification
[0170] According to the hygromycin resistance gene sequence (Hyg) in the pCAMBIA-1301 plasmid, a pair of gene-specific primers (upstream primer Hyg-F, plus restriction enzyme site BstEII (GGT TACC) and protective bases were designed at the beginning and end respectively , downstream primer Hyg-R, plus restriction site BstEII (GGT GACC) and protective bases), pCAMBIA-1301 plasmid as a template, high-fidelity Taq polymerase for PCR amplification.
[0171] Hyg-F: A TCAGTTAGCCTCCCCCATCTCC (SEQ ID NO: 12)
[0172] Hyg-R: A ATATGAAAAAGCCTGAACTCA (SEQ ID NO: 13)
[0173] PCR system: 25μl
[0174] wxya 2 O 11.3 μl
[0175] 2*Primer Star Mix 12.5μl
[0176] pCAMBIA-1301 plasmid 0.2μl
[0177] Primer Hyg-F 0.5μl
[0178] Primer Hyg-R 0.5μl
[0179] PC...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com