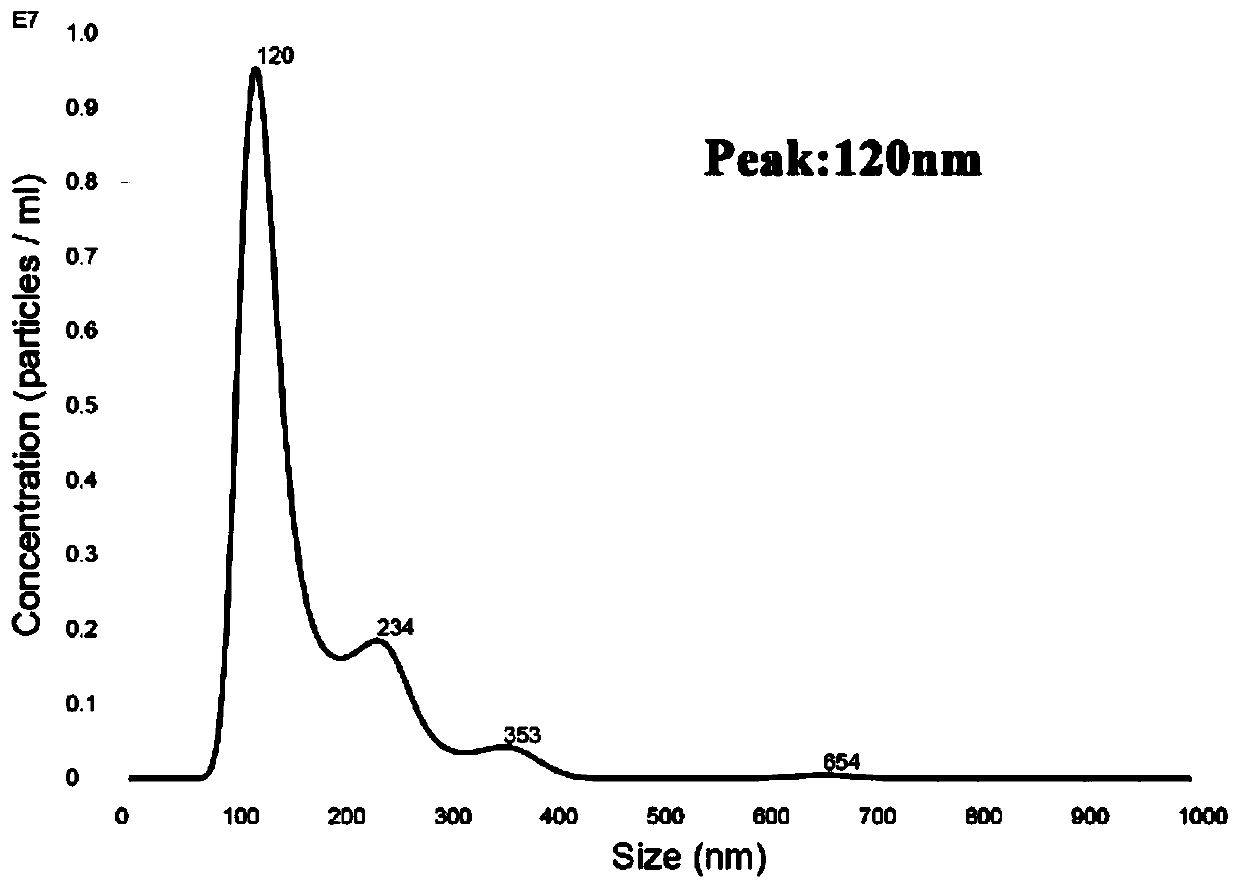
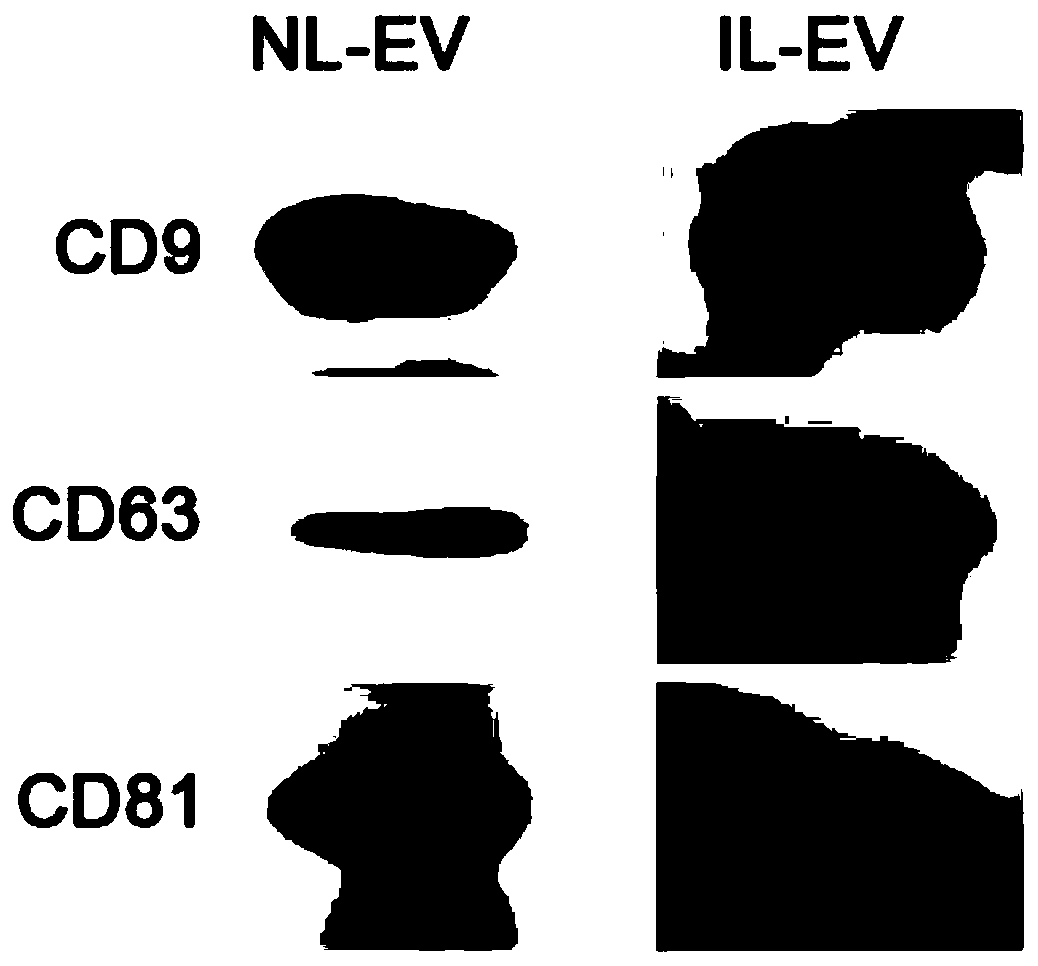
Method for detecting schistosoma japonicum infection by using host exosome miRNA-142a-3p
A technology of schistosomiasis and exosomes, applied in the direction of DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problem of lack of rapid, sensitive, specific and stable detection methods for Schistosoma japonicum, low sensitivity, Insufficient specificity and other issues, to achieve good clinical application value, high sensitivity, and reduce the effect of patient trauma
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1 A diagnostic kit for Schistosoma japonicum infection
[0051] 1. Composition
[0052] Amplification primers for serum exosome miR-142a-3p, internal reference gene U6 amplification primers and q-PCR reagents;
[0053] Among them, the upstream primer nucleotide of serum exosomal miR-142a-3p is shown in SEQ ID NO: 2 (CTGTAGTGTTTCCTACTTTATG), and the downstream primer is provided in the commercially available microRNA quantitative (qRT-PCR) (tailing method) kit Universal primers;
[0054] The nucleotide sequence of the internal reference gene U6 amplification primer is shown in SEQ ID NO: 3-4,
[0055] SEQ ID NO: 3: GGAACGATACAGAGAAGATTAGC,
[0056] SEQ ID NO: 4: TGGAACGCTTCACGAATTTGCG;
[0057] The reagents for q-PCR are Premix Ex Taq TM , ROX Dye (50×) and ddH 2 O.
[0058] 2. How to use
[0059] 1. Extract exosomes from serum samples to be tested
[0060] Use a commercially available serum exosome extraction kit.
[0061] 2. Extract total exosome RN...
Embodiment 2
[0082] Embodiment 2 detection sensitivity effect analysis
[0083] 1. Experimental method
[0084] The kit in Example 1 was used to detect serum exosomal miR-142a-3p from healthy people from schistosomiasis-endemic areas and schistosomiasis-infected groups.
[0085] 2. Experimental results
[0086] The expression of exosomal miR-142a-3p in schistosomiasis-infected people was higher than that in healthy people, which was more than 2 times higher.
Embodiment 3
[0087] Embodiment 3 mouse model experiment verification sensitivity
[0088] 1. Experimental method
[0089] The schistosomiasis-infected mouse model was routinely constructed, and the sera of mice in the uninfected control group and the infected group were collected, and exosomes were extracted. The kit of claim 2 is used to detect serum exosomal miR-142a-3p of mice in the uninfected control group and the infected group.
[0090] 2. Experimental results
[0091] The serum exosomal miR-142a-3p expression level of mice in the infected group was higher than that in the uninfected control group, which was more than 2 times higher.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com